[English] 日本語
 Yorodumi
Yorodumi- EMDB-23081: High resolution cryo EM analysis of HPV16 identifies minor struct... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-23081 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
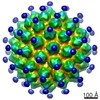
| Title | High resolution cryo EM analysis of HPV16 identifies minor structural protein L2 and describes capsid flexibility | |||||||||
 Map data Map data | HPV16 quasivirus capsid. Recombined map from pentavalent and hexavalent subvolume refinements. | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | HPV16 / quasivirus / L1 / capsomer / subparticle / VIRUS | |||||||||
| Function / homology |  Function and homology information Function and homology informationT=7 icosahedral viral capsid / endocytosis involved in viral entry into host cell / virion attachment to host cell / host cell nucleus / structural molecule activity Similarity search - Function | |||||||||
| Biological species |  Human papillomavirus type 16 Human papillomavirus type 16 | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.1 Å | |||||||||
 Authors Authors | Hartmann SR / Goetschius DJ | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Sci Rep / Year: 2021 Journal: Sci Rep / Year: 2021Title: High resolution cryo EM analysis of HPV16 identifies minor structural protein L2 and describes capsid flexibility. Authors: Daniel J Goetschius / Samantha R Hartmann / Suriyasri Subramanian / Carol M Bator / Neil D Christensen / Susan L Hafenstein /  Abstract: Human papillomavirus (HPV) is a significant health burden and leading cause of virus-induced cancers. HPV is epitheliotropic and its replication is tightly associated with terminal keratinocyte ...Human papillomavirus (HPV) is a significant health burden and leading cause of virus-induced cancers. HPV is epitheliotropic and its replication is tightly associated with terminal keratinocyte differentiation making production and purification of high titer virus preparations for research problematic, therefore alternative HPV production methods have been developed for virological and structural studies. In this study we use HPV16 quasivirus, composed of HPV16 L1/L2 capsid proteins with a packaged cottontail rabbit papillomavirus genome. We have achieved the first high resolution, 3.1 Å, structure of HPV16 by using a local subvolume refinement approach. The high resolution enabled us to build L1 unambiguously and identify L2 protein strands. The L2 density is incorporated adjacent to conserved L1 residues on the interior of the capsid. Further interpretation with our own software for Icosahedral Subvolume Extraction and Correlated Classification revealed flexibility, on the whole-particle level through diameter analysis and local movement with inter-capsomer analysis. Inter-capsomer expansion or contraction, governed by the connecting arms, showed no bias in the magnitude or direction of capsomer movement. We propose that papillomavirus capsids are dynamic and capsomers move as rigid bodies connected by flexible linkers. The resulting virus structure will provide a framework for continuing biochemical, genetic and biophysical research for papillomaviruses. Furthermore, our approach has allowed insight into the resolution barrier that has previously been a limitation in papillomavirus structural studies. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_23081.map.gz emd_23081.map.gz | 696.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-23081-v30.xml emd-23081-v30.xml emd-23081.xml emd-23081.xml | 16.8 KB 16.8 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_23081.png emd_23081.png | 225.5 KB | ||
| Filedesc metadata |  emd-23081.cif.gz emd-23081.cif.gz | 5.7 KB | ||
| Others |  emd_23081_additional_1.map.gz emd_23081_additional_1.map.gz emd_23081_additional_2.map.gz emd_23081_additional_2.map.gz emd_23081_additional_3.map.gz emd_23081_additional_3.map.gz | 96.2 MB 96.1 MB 1.2 GB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-23081 http://ftp.pdbj.org/pub/emdb/structures/EMD-23081 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-23081 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-23081 | HTTPS FTP |
-Related structure data
| Related structure data |  7kzfMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_23081.map.gz / Format: CCP4 / Size: 824 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_23081.map.gz / Format: CCP4 / Size: 824 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | HPV16 quasivirus capsid. Recombined map from pentavalent and hexavalent subvolume refinements. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.1 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
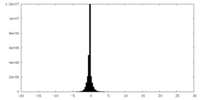
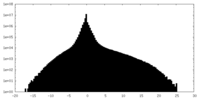
-Additional map: HPV16 quasivirus pentavalent capsomer subvolume refinement.
| File | emd_23081_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | HPV16 quasivirus pentavalent capsomer subvolume refinement. | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
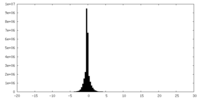
-Additional map: HPV16 quasivirus hexavalent capsomer subvolume refinement.
| File | emd_23081_additional_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | HPV16 quasivirus hexavalent capsomer subvolume refinement. | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
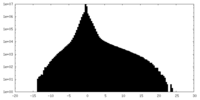
-Additional map: HPV16 quasivirus icosahedral refinement from which pentavalent and...
| File | emd_23081_additional_3.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | HPV16 quasivirus icosahedral refinement from which pentavalent and hexavalent subvolumes were extracted. | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Human papillomavirus type 16
| Entire | Name:  Human papillomavirus type 16 Human papillomavirus type 16 |
|---|---|
| Components |
|
-Supramolecule #1: Human papillomavirus type 16
| Supramolecule | Name: Human papillomavirus type 16 / type: virus / ID: 1 / Parent: 0 / Macromolecule list: all / NCBI-ID: 333760 / Sci species name: Human papillomavirus type 16 / Virus type: VIRUS-LIKE PARTICLE / Virus isolate: STRAIN / Virus enveloped: No / Virus empty: No |
|---|
-Macromolecule #1: Major capsid protein L1
| Macromolecule | Name: Major capsid protein L1 / type: protein_or_peptide / ID: 1 / Number of copies: 6 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Human papillomavirus type 16 Human papillomavirus type 16 |
| Molecular weight | Theoretical: 56.098617 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MSLWLPSEAT VYLPPVPVSK VVSTDEYVAR TNIYYHAGTS RLLAVGHPYF PIKKPNNNKI LVPKVSGLQY RVFRIHLPDP NKFGFPDTS FYNPDTQRLV WACVGVEVGR GQPLGVGISG HPLLNKLDDT ENASAYAANA GVDNRECISM DYKQTQLCLI G CKPPIGEH ...String: MSLWLPSEAT VYLPPVPVSK VVSTDEYVAR TNIYYHAGTS RLLAVGHPYF PIKKPNNNKI LVPKVSGLQY RVFRIHLPDP NKFGFPDTS FYNPDTQRLV WACVGVEVGR GQPLGVGISG HPLLNKLDDT ENASAYAANA GVDNRECISM DYKQTQLCLI G CKPPIGEH WGKGSPCTNV AVNPGDCPPL ELINTVIQDG DMVDTGFGAM DFTTLQANKS EVPLDICTSI CKYPDYIKMV SE PYGDSLF FYLRREQMFV RHLFNRAGAV GENVPDDLYI KGSGSTANLA SSNYFPTPSG SMVTSDAQIF NKPYWLQRAQ GHN NGICWG NQLFVTVVDT TRSTNMSLCA AISTSETTYK NTNFKEYLRH GEEYDLQFIF QLCKITLTAD VMTYIHSMNS TILE DWNFG LQPPPGGTLE DTYRFVTSQA IACQKHTPPA PKEDPLKKYT FWEVNLKEKF SADLDQFPLG RKFLLQAGLK AKPKF TLGK RKATPTTSST STTAKRKKR UniProtKB: Major capsid protein L1 |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 / Component - Concentration: 1.0 X / Component - Name: PBS |
|---|---|
| Grid | Pretreatment - Type: GLOW DISCHARGE / Details: unspecified |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON III (4k x 4k) / Detector mode: COUNTING / Number grids imaged: 1 / Number real images: 10143 / Average electron dose: 60.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 0.01 mm |
| Sample stage | Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)