+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-9920 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
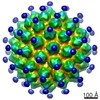
| Title | The cryo-em structure of HPV18 VLP | |||||||||
 Map data Map data | The cryo-em structure of HPV18 VLP | |||||||||
 Sample Sample |
| |||||||||
| Biological species |   Human papillomavirus Human papillomavirus | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 11.5 Å | |||||||||
 Authors Authors | Li S / Zheng Q | |||||||||
 Citation Citation |  Journal: Eur J Pharm Biopharm / Year: 2019 Journal: Eur J Pharm Biopharm / Year: 2019Title: Altered antigenicity and immunogenicity of human papillomavirus virus-like particles in the presence of thimerosal. Authors: Siyi Chen / Xiaofen Huang / Yike Li / Xin Wang / Huirong Pan / Zhijie Lin / Qingbing Zheng / Shaowei Li / Jun Zhang / Ningshao Xia / Qinjian Zhao /  Abstract: Thimerosal has been widely used as a preservative in human vaccines for decades. Thimerosal, a thiol capping agent with ethyl mercury being the active degradant, could have impacts on the vaccine ...Thimerosal has been widely used as a preservative in human vaccines for decades. Thimerosal, a thiol capping agent with ethyl mercury being the active degradant, could have impacts on the vaccine potency due to potential thiol modification. The effects on the antigenicity and immunogenicity of human papillomavirus (HPV) virus-like particles (VLPs) in the presence of thimerosal was studied. In general, reduced binding activity was observed between HPV antigens and monoclonal antibodies (mAbs) upon thimerosal treatment, accompanied by reduced protein conformational stability. The immunogenicity of a pentavalent vaccine formulation (HPV6, HPV11, HPV16, HPV18 and hepatitis E virus) with or without thimerosal was studied in mice. The functional antibody titres, as well as the binding titres, were determined, showing a substantial decrease for vaccine formulations containing thimerosal for HPV16/18. Similarly, epitope-specific competition assays using specific and functional mAbs as tracers also showed a significant reduction in immunogenicity for HPV16/18 in the presence of thimerosal. Structural alterations in the capsid protein for HPV18 were observed with cryo-electron microscopy and 3-dimensional reconstruction in the comparative structural analysis. The results should alert scientists in formulation development field on the choice for vaccine preservatives, in particular for thiol-containing antigens. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_9920.map.gz emd_9920.map.gz | 714.8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-9920-v30.xml emd-9920-v30.xml emd-9920.xml emd-9920.xml | 8.6 KB 8.6 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_9920.png emd_9920.png | 203.8 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-9920 http://ftp.pdbj.org/pub/emdb/structures/EMD-9920 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-9920 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-9920 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_9920.map.gz / Format: CCP4 / Size: 1.9 GB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_9920.map.gz / Format: CCP4 / Size: 1.9 GB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | The cryo-em structure of HPV18 VLP | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.128 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Human papillomavirus
| Entire | Name:   Human papillomavirus Human papillomavirus |
|---|---|
| Components |
|
-Supramolecule #1: Human papillomavirus
| Supramolecule | Name: Human papillomavirus / type: virus / ID: 1 / Parent: 0 / NCBI-ID: 10566 / Sci species name: Human papillomavirus / Virus type: VIRUS-LIKE PARTICLE / Virus isolate: OTHER / Virus enveloped: No / Virus empty: Yes |
|---|---|
| Host system | Organism: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 2.0 mg/mL |
|---|---|
| Buffer | pH: 7.4 |
| Grid | Model: Quantifoil R2/2 / Material: COPPER / Mesh: 200 / Pretreatment - Type: GLOW DISCHARGE |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TECNAI F30 |
|---|---|
| Image recording | Film or detector model: FEI FALCON III (4k x 4k) / Detector mode: INTEGRATING / Average electron dose: 25.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Sample stage | Specimen holder model: GATAN 626 SINGLE TILT LIQUID NITROGEN CRYO TRANSFER HOLDER Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Tecnai F30 / Image courtesy: FEI Company |
- Image processing
Image processing
| Final reconstruction | Applied symmetry - Point group: I (icosahedral) / Resolution.type: BY AUTHOR / Resolution: 11.5 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 901 |
|---|---|
| Initial angle assignment | Type: RANDOM ASSIGNMENT |
| Final angle assignment | Type: ANGULAR RECONSTITUTION |
 Movie
Movie Controller
Controller

















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)





















