[English] 日本語
 Yorodumi
Yorodumi- EMDB-30781: The cryo-EM structure of human papillomavirus type 58 pseudovirus -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-30781 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
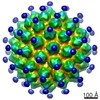
| Title | The cryo-EM structure of human papillomavirus type 58 pseudovirus | |||||||||
 Map data Map data | The Cryo-EM structure of human papillomavirus type pseudovirus | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | particles / VIRUS | |||||||||
| Function / homology |  Function and homology information Function and homology informationT=7 icosahedral viral capsid / endocytosis involved in viral entry into host cell / virion attachment to host cell / host cell nucleus / structural molecule activity Similarity search - Function | |||||||||
| Biological species |   Human papillomavirus type 58 Human papillomavirus type 58 | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 4.11 Å | |||||||||
 Authors Authors | He MZ / Chi X | |||||||||
| Funding support |  China, 1 items China, 1 items
| |||||||||
 Citation Citation |  Journal: J Virol / Year: 2021 Journal: J Virol / Year: 2021Title: Structural basis for the shared neutralization mechanism of three classes of human papillomavirus type 58 antibodies with disparate modes of binding. Authors: Maozhou He / Xin Chi / Zhenghui Zha / Yunbing Li / Jie Chen / Yang Huang / Shiwen Huang / Miao Yu / Zhiping Wang / Shuo Song / Xinlin Liu / Shuangping Wei / Zekai Li / Tingting Li / Yingbin ...Authors: Maozhou He / Xin Chi / Zhenghui Zha / Yunbing Li / Jie Chen / Yang Huang / Shiwen Huang / Miao Yu / Zhiping Wang / Shuo Song / Xinlin Liu / Shuangping Wei / Zekai Li / Tingting Li / Yingbin Wang / Hai Yu / Qinjian Zhao / Jun Zhang / Qingbing Zheng / Ying Gu / Shaowei Li / Ningshao Xia /  Abstract: Human papillomavirus type 58 (HPV58) is associated with cervical cancer and poses a significant health burden worldwide. Although the commercial 9-valent HPV vaccine covers HPV58, the structural and ...Human papillomavirus type 58 (HPV58) is associated with cervical cancer and poses a significant health burden worldwide. Although the commercial 9-valent HPV vaccine covers HPV58, the structural and molecular-level neutralization sites of the HPV58 complete virion are not fully understood. Here, we report the high-resolution (∼3.5 Å) structure of the complete HPV58 pseudovirus (PsV58) using cryo-electron microscopy (cryo-EM). Three representative neutralizing monoclonal antibodies (nAbs 5G9, 2H3 and A4B4) were selected through clustering from a nAb panel against HPV58. Bypassing the steric hindrance and symmetry-mismatch in the HPV Fab-capsid immune-complex, we present three different neutralizing epitopes in the PsV58, and show that, despite differences in binding, these nAbs share a common neutralization mechanism. These results offer insight into HPV58 genotype specificity and broaden our understanding of HPV58 neutralization sites for antiviral research. Cervical cancer primarily results from persistent infection with high-risk types of human papillomavirus (HPV). HPV type 58 (HPV58) is an important causative agent, especially within Asia. Despite this, we still have limited data pertaining to the structural and neutralizing epitopes of HPV58, and this encumbers our in-depth understanding of the virus mode of infection. Here, we show that representative nAbs (5G9, 10B11, 2H3, 5H2 and A4B4) from three different groups share a common neutralization mechanism that appears to prohibit the virus from associating with the extracellular matrix and cell surface. Furthermore, we identify that the nAbs engage via three different binding patterns: top-center binding (5G9 and 10B11), top-fringe binding (2H3 and 5H2), and fringe binding (A4B4). Our work shows that, despite differences in the pattern in binding, nAbs against HPV58 share a common neutralization mechanism. These results provide new insight into the understanding of HPV58 infection. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_30781.map.gz emd_30781.map.gz | 1.3 GB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-30781-v30.xml emd-30781-v30.xml emd-30781.xml emd-30781.xml | 8.8 KB 8.8 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_30781.png emd_30781.png | 288.1 KB | ||
| Filedesc metadata |  emd-30781.cif.gz emd-30781.cif.gz | 5.1 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-30781 http://ftp.pdbj.org/pub/emdb/structures/EMD-30781 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-30781 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-30781 | HTTPS FTP |
-Related structure data
| Related structure data |  7dn5MC  7dnhC  7dnkC  7dnlC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_30781.map.gz / Format: CCP4 / Size: 1.4 GB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_30781.map.gz / Format: CCP4 / Size: 1.4 GB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | The Cryo-EM structure of human papillomavirus type pseudovirus | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.128 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Human papillomavirus type 58
| Entire | Name:   Human papillomavirus type 58 Human papillomavirus type 58 |
|---|---|
| Components |
|
-Supramolecule #1: Human papillomavirus type 58
| Supramolecule | Name: Human papillomavirus type 58 / type: virus / ID: 1 / Parent: 0 / Macromolecule list: all / NCBI-ID: 10598 / Sci species name: Human papillomavirus type 58 / Virus type: VIRION / Virus isolate: SPECIES / Virus enveloped: No / Virus empty: No |
|---|
-Macromolecule #1: Major capsid protein L1
| Macromolecule | Name: Major capsid protein L1 / type: protein_or_peptide / ID: 1 / Number of copies: 6 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Human papillomavirus type 58 Human papillomavirus type 58 |
| Molecular weight | Theoretical: 59.103199 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MVLILCCTLA ILFCVADVNV FHIFLQMSVW RPSEATVYLP PVPVSKVVST DEYVSRTSIY YYAGSSRLLA VGNPYFSIKS PNNNKKVLV PKVSGLQYRV FRVRLPDPNK FGFPDTSFYN PDTQRLVWAC VGLEIGRGQP LGVGVSGHPY LNKFDDTETS N RYPAQPGS ...String: MVLILCCTLA ILFCVADVNV FHIFLQMSVW RPSEATVYLP PVPVSKVVST DEYVSRTSIY YYAGSSRLLA VGNPYFSIKS PNNNKKVLV PKVSGLQYRV FRVRLPDPNK FGFPDTSFYN PDTQRLVWAC VGLEIGRGQP LGVGVSGHPY LNKFDDTETS N RYPAQPGS DNRECLSMDY KQTQLCLIGC KPPTGEHWGK GVACNNNAAA TDCPPLELFN SIIEDGDMVD TGFGCMDFGT LQ ANKSDVP IDICNSTCKY PDYLKMASEP YGDSLFFFLR REQMFVRHFF NRAGKLGEAV PDDLYIKGSG NTAVIQSSAF FPT PSGSIV TSESQLFNKP YWLQRAQGHN NGICWGNQLF VTVVDTTRST NMTLCTEVTK EGTYKNDNFK EYVRHVEEYD LQFV FQLCK ITLTAEIMTY IHTMDSNILE DWQFGLTPPP SASLQDTYRF VTSQAITCQK TAPPKEKEDP LNKYTFWEVN LKEKF SADL DQFPLGRKFL LQSGLKAKPR LKRSAPTTRA PSTKRKKVKK UniProtKB: Major capsid protein L1 |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TECNAI F30 |
|---|---|
| Image recording | Film or detector model: FEI FALCON II (4k x 4k) / Detector mode: INTEGRATING / Average electron dose: 30.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Tecnai F30 / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: OTHER |
|---|---|
| Final reconstruction | Applied symmetry - Point group: I (icosahedral) / Resolution.type: BY AUTHOR / Resolution: 4.11 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 13458 |
| Initial angle assignment | Type: COMMON LINE |
| Final angle assignment | Type: MAXIMUM LIKELIHOOD |
 Movie
Movie Controller
Controller

























 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)





















