[English] 日本語
 Yorodumi
Yorodumi- EMDB-11861: Helical reconstruction of nitrite oxidoreductase from anammox bacteria -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-11861 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Helical reconstruction of nitrite oxidoreductase from anammox bacteria | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
| Function / homology |  Function and homology information Function and homology informationnitrate reductase / anammoxosome / cell envelope / anaerobic respiration / 3 iron, 4 sulfur cluster binding / 4 iron, 4 sulfur cluster binding / oxidoreductase activity / electron transfer activity / periplasmic space / heme binding ...nitrate reductase / anammoxosome / cell envelope / anaerobic respiration / 3 iron, 4 sulfur cluster binding / 4 iron, 4 sulfur cluster binding / oxidoreductase activity / electron transfer activity / periplasmic space / heme binding / metal ion binding / membrane Similarity search - Function | |||||||||
| Biological species |  Candidatus Kuenenia stuttgartiensis (bacteria) Candidatus Kuenenia stuttgartiensis (bacteria) | |||||||||
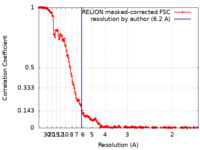
| Method | helical reconstruction / cryo EM / Resolution: 6.2 Å | |||||||||
 Authors Authors | Parey K | |||||||||
 Citation Citation |  Journal: Nat Microbiol / Year: 2021 Journal: Nat Microbiol / Year: 2021Title: Structural and functional characterization of the intracellular filament-forming nitrite oxidoreductase multiprotein complex. Authors: Tadeo Moreno Chicano / Lea Dietrich / Naomi M de Almeida / Mohd Akram / Elisabeth Hartmann / Franziska Leidreiter / Daniel Leopoldus / Melanie Mueller / Ricardo Sánchez / Guylaine H L ...Authors: Tadeo Moreno Chicano / Lea Dietrich / Naomi M de Almeida / Mohd Akram / Elisabeth Hartmann / Franziska Leidreiter / Daniel Leopoldus / Melanie Mueller / Ricardo Sánchez / Guylaine H L Nuijten / Joachim Reimann / Kerstin-Anikó Seifert / Ilme Schlichting / Laura van Niftrik / Mike S M Jetten / Andreas Dietl / Boran Kartal / Kristian Parey / Thomas R M Barends /   Abstract: Nitrate is an abundant nutrient and electron acceptor throughout Earth's biosphere. Virtually all nitrate in nature is produced by the oxidation of nitrite by the nitrite oxidoreductase (NXR) ...Nitrate is an abundant nutrient and electron acceptor throughout Earth's biosphere. Virtually all nitrate in nature is produced by the oxidation of nitrite by the nitrite oxidoreductase (NXR) multiprotein complex. NXR is a crucial enzyme in the global biological nitrogen cycle, and is found in nitrite-oxidizing bacteria (including comammox organisms), which generate the bulk of the nitrate in the environment, and in anaerobic ammonium-oxidizing (anammox) bacteria which produce half of the dinitrogen gas in our atmosphere. However, despite its central role in biology and decades of intense study, no structural information on NXR is available. Here, we present a structural and biochemical analysis of the NXR from the anammox bacterium Kuenenia stuttgartiensis, integrating X-ray crystallography, cryo-electron tomography, helical reconstruction cryo-electron microscopy, interaction and reconstitution studies and enzyme kinetics. We find that NXR catalyses both nitrite oxidation and nitrate reduction, and show that in the cell, NXR is arranged in tubules several hundred nanometres long. We reveal the tubule architecture and show that tubule formation is induced by a previously unidentified, haem-containing subunit, NXR-T. The results also reveal unexpected features in the active site of the enzyme, an unusual cofactor coordination in the protein's electron transport chain, and elucidate the electron transfer pathways within the complex. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_11861.map.gz emd_11861.map.gz | 75.8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-11861-v30.xml emd-11861-v30.xml emd-11861.xml emd-11861.xml | 22.3 KB 22.3 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_11861_fsc.xml emd_11861_fsc.xml | 17.2 KB | Display |  FSC data file FSC data file |
| Images |  emd_11861.png emd_11861.png | 224.3 KB | ||
| Masks |  emd_11861_msk_1.map emd_11861_msk_1.map | 421.9 MB |  Mask map Mask map | |
| Others |  emd_11861_additional_1.map.gz emd_11861_additional_1.map.gz emd_11861_half_map_1.map.gz emd_11861_half_map_1.map.gz emd_11861_half_map_2.map.gz emd_11861_half_map_2.map.gz | 393.7 MB 337.4 MB 337.4 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-11861 http://ftp.pdbj.org/pub/emdb/structures/EMD-11861 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-11861 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-11861 | HTTPS FTP |
-Related structure data
| Related structure data | C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_11861.map.gz / Format: CCP4 / Size: 81.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_11861.map.gz / Format: CCP4 / Size: 81.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. generated in cubic-lattice coordinate | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.837 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Mask #1
| File |  emd_11861_msk_1.map emd_11861_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: #1
| File | emd_11861_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_11861_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_11861_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Nitrite oxidoreductase
| Entire | Name: Nitrite oxidoreductase |
|---|---|
| Components |
|
-Supramolecule #1: Nitrite oxidoreductase
| Supramolecule | Name: Nitrite oxidoreductase / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Candidatus Kuenenia stuttgartiensis (bacteria) Candidatus Kuenenia stuttgartiensis (bacteria) |
| Molecular weight | Theoretical: 230 kDa/nm |
-Macromolecule #1: Nitrite oxidoreductase subunit A
| Macromolecule | Name: Nitrite oxidoreductase subunit A / type: protein_or_peptide / ID: 1 / Enantiomer: LEVO / EC number: nitrate reductase |
|---|---|
| Source (natural) | Organism:  Candidatus Kuenenia stuttgartiensis (bacteria) Candidatus Kuenenia stuttgartiensis (bacteria) |
| Sequence | String: MKLTRRAFLQ VAGATGATLT LAKNAMAFRL LKPAVVVDNP LDTYPDRRWE SVYRDQYQYD RTFTYCCSP NDTHACRIRA FVRNNVMMRV EQNYDHQNYS DLYGNKATRN WNPRMCLKGY T FHRRVYGP YRLRYPLIRK GWKRWADDGF PELTPENKTK YMFDNRGNDE ...String: MKLTRRAFLQ VAGATGATLT LAKNAMAFRL LKPAVVVDNP LDTYPDRRWE SVYRDQYQYD RTFTYCCSP NDTHACRIRA FVRNNVMMRV EQNYDHQNYS DLYGNKATRN WNPRMCLKGY T FHRRVYGP YRLRYPLIRK GWKRWADDGF PELTPENKTK YMFDNRGNDE LLRASWDEAF TY ASKGIIH ITKKYSGPEG AQKLIDQGYP KEMVDRMQGA GTRTFKGRGG MGLLGVIGKY GMY RFNNCL AIVDAHNRGV GPDQALGGRN WSNYTWHGDQ APGHPFSHGL QTSDVDMNDV RFSK LLIQT GKNLIENKMP EAHWVTEVME RGGKIVVITP EYSPSAQKAD YWIPIRNNTD TALFL GITK ILIDNKWYDA DYVKKFTDFP LLIRTDTLKR VSPKDIIPNY KLQDISDGPS YHIQGL KDE QREIIGDFVV WDAKSKGPKA ITRDDVGETL VKKGIDPVLE GSFKLKTIDG KEIEVMT LL EMYKIHLRDY DIDSVVSMTN SPKDLIERLA KDIATIKPVA IHYGEGVNHY FHATLMNR S YYLPVMLTGN VGYFGSGSHT WAGNYKAGNF QASKWSGPGF YGWVAEDVFK PNLDPYASA KDLNIKGRAL DEEVAYWNHS ERPLIVNTPK YGRKVFTGKT HMPSPTKVLW FTNVNLINNA KHVYQMLKN VNPNIEQIMS TDIEITGSIE YADFAFPANS WVEFQEFEIT NSCSNPFIQI W GKTGITPV YESKDDVKIL AGMASKLGEL LRDKRFEDNW KFAIEGRASV YINRLLDGST TM KGYTCED ILNGKYGEPG VAMLLFRTYP RHPFWEQVHE SLPFYTPTGR LQAYNDEPEI IEY GENFIV HREGPEATPY LPNAIVSTNP YIRPDDYGIP ENAEYWEDRT VRNIKKSWEE TKKT KNFLW EKGYHFYCVT PKSRHTVHSQ WAVTDWNFIW NNNFGDPYRM DKRMPGVGEH QIHIH PQAA RDLGIEDGDY VYVDANPADR PYEGWKPNDS FYKVSRLMLR AKYNPAYPYN CTMMKH SAW ISSDKTVQAH ETRPDGRALS PSGYQSSFRY GSQQSITRDW SMPMHQLDSL FHKAKIG MK FIFGFEADNH CINTVPKETL VKITKAENGG MGGKGVWDPV KTGYTAGNEN DFMKKFLN G ELIKVDA |
-Macromolecule #2: Nitrite oxidoreductase subunit B
| Macromolecule | Name: Nitrite oxidoreductase subunit B / type: protein_or_peptide / ID: 2 / Enantiomer: LEVO / EC number: nitrate reductase |
|---|---|
| Source (natural) | Organism:  Candidatus Kuenenia stuttgartiensis (bacteria) Candidatus Kuenenia stuttgartiensis (bacteria) |
| Sequence | String: MTLVHNWHLG RRMEYPYFES RPKHQFAAVF NINRCIACQT CTMACKSTWT FNKGQEFMWW NNVETKPYG GFPQSWDVKT LKLIDSPDNI WYTDDKDKET SQYGTGAPYG TYEGDTIFEV A KKKNINQW AVGYIPEDKE WRSPNFGEDT AKSSNQPGEY STLPEHSRWF ...String: MTLVHNWHLG RRMEYPYFES RPKHQFAAVF NINRCIACQT CTMACKSTWT FNKGQEFMWW NNVETKPYG GFPQSWDVKT LKLIDSPDNI WYTDDKDKET SQYGTGAPYG TYEGDTIFEV A KKKNINQW AVGYIPEDKE WRSPNFGEDT AKSSNQPGEY STLPEHSRWF FYLQRICNHC TY PGCLAAC PRKAIYKRKE DGIVLIDQKR CRGYRKCVEQ CPYKKPMYRG LTRVSEKCIA CYP RIEGRD SLTDGRPMET RCMSACVGQI RLQGFLDDNP KNPITWLIRH QKIALPLYPQ FGTE PNIYY IPPRWAPRAY LRQMFGPGVD EAIEKFMVPS RELLAVMSLF RMTQTIVYEY KIEEG PKVF ETEIHGKKFT MYNDTVIGFG EDGKEVVRTT VEEPIHIRPD KHYNSI |
-Macromolecule #3: Nitrite oxidoreductase subunit C
| Macromolecule | Name: Nitrite oxidoreductase subunit C / type: protein_or_peptide / ID: 3 / Enantiomer: LEVO / EC number: nitrate reductase |
|---|---|
| Source (natural) | Organism:  Candidatus Kuenenia stuttgartiensis (bacteria) Candidatus Kuenenia stuttgartiensis (bacteria) |
| Sequence | String: MKKFYRLLGS SSVALLGCLF LSVALCIAEE AEGVKGVAEE ELTPAKEVLN VKYMQIDVPA HITVGALEG AFKNAEGVQV KLQKQDKAFP NGGGSVNSAE IKAIHDGITI YFQVIWDDAT D NKQAIATQ EFRDGAALMF PLGKITISPE EPFSPRMGDR QKPVNLWHWK ...String: MKKFYRLLGS SSVALLGCLF LSVALCIAEE AEGVKGVAEE ELTPAKEVLN VKYMQIDVPA HITVGALEG AFKNAEGVQV KLQKQDKAFP NGGGSVNSAE IKAIHDGITI YFQVIWDDAT D NKQAIATQ EFRDGAALMF PLGKITISPE EPFSPRMGDR QKPVNLWHWK ADWEADLLAT GG IEECPAR YPNMHDDFST NPHSVNYHKG VIQSAAELSG GYAAHNLLSL PRGRAVEDLN AEG FGTLTS QDHQDVDGCS KFENKKWTVV FCRSLNTGDP LDVQFVPGES TYFNMAVWNG DRED RNGQK NISIQWHPLS LERIAWQ |
-Macromolecule #4: Conserved hypothetical (Monoheme) protein
| Macromolecule | Name: Conserved hypothetical (Monoheme) protein / type: protein_or_peptide / ID: 4 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Candidatus Kuenenia stuttgartiensis (bacteria) Candidatus Kuenenia stuttgartiensis (bacteria) |
| Sequence | String: MGMIKMKYVV FGVLALIMLG IGNSMFTHKA MGSSGKAAEH EGVIEPTQEL MKDIEKRLTN MLSGILENN LKYVANEAGA VVDQSYKISE FFFPFDPKKN EWFERAGIDP KDAGKITKLR E EFALFQSG IVYKALNVRK MAMEGSQEET LKAFADLIEK TCFACHKKNR DWLFDQPGSH GP GR |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | helical reconstruction |
| Aggregation state | helical array |
- Sample preparation
Sample preparation
| Concentration | 1.5 mg/mL | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.5 Component:
| ||||||||||||
| Grid | Model: C-flat-1.2/1.3 / Material: COPPER / Mesh: 300 / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE | ||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 70 % / Chamber temperature: 283 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Specialist optics | Energy filter - Name: GIF Bioquantum / Energy filter - Slit width: 30 eV |
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Detector mode: COUNTING / Number real images: 1753 / Average exposure time: 2.0 sec. / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: -3.0 µm / Nominal defocus min: -2.0 µm / Nominal magnification: 105000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller























 X (Sec.)
X (Sec.) Y (Row.)
Y (Row.) Z (Col.)
Z (Col.)