[English] 日本語
 Yorodumi
Yorodumi- EMDB-22615: Cryo-EM structure of rabbit RyR1 in the presence of Mg2+ and AMP-... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-22615 | |||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of rabbit RyR1 in the presence of Mg2+ and AMP-PCP in nanodisc | |||||||||||||||||||||
 Map data Map data | Post-processed full map | |||||||||||||||||||||
 Sample Sample |
| |||||||||||||||||||||
 Keywords Keywords | Ryanodine Receptor / RyR1 / Intracellular Calcium channel / Mg2+ / Inhibition / Excitation-Contraction coupling / TRANSPORT PROTEIN | |||||||||||||||||||||
| Biological species |  | |||||||||||||||||||||
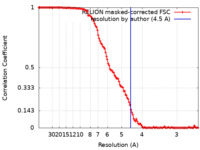
| Method | single particle reconstruction / cryo EM / Resolution: 4.5 Å | |||||||||||||||||||||
 Authors Authors | Nayak AR / Samso M | |||||||||||||||||||||
| Funding support |  United States, 6 items United States, 6 items
| |||||||||||||||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2024 Journal: Nat Commun / Year: 2024Title: Interplay between Mg2+ and Ca2+ at multiple sites of the ryanodine receptor Authors: Nayak AR / Rangubpit W / Will AH / Hu Y / Castro-Hartmann P / Lobo JJ / Dryden K / Lamb GD / Sompornpisut P / Samso M | |||||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_22615.map.gz emd_22615.map.gz | 17.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-22615-v30.xml emd-22615-v30.xml emd-22615.xml emd-22615.xml | 35.4 KB 35.4 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_22615_fsc.xml emd_22615_fsc.xml | 12.2 KB | Display |  FSC data file FSC data file |
| Images |  emd_22615.png emd_22615.png | 144.3 KB | ||
| Masks |  emd_22615_msk_1.map emd_22615_msk_1.map | 166.4 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-22615.cif.gz emd-22615.cif.gz | 9.3 KB | ||
| Others |  emd_22615_additional_1.map.gz emd_22615_additional_1.map.gz emd_22615_additional_2.map.gz emd_22615_additional_2.map.gz emd_22615_half_map_1.map.gz emd_22615_half_map_1.map.gz emd_22615_half_map_2.map.gz emd_22615_half_map_2.map.gz | 126.5 MB 126.6 MB 15.8 MB 15.8 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-22615 http://ftp.pdbj.org/pub/emdb/structures/EMD-22615 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-22615 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-22615 | HTTPS FTP |
-Related structure data
| Related structure data |  7k0sMC  7umzC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_22615.map.gz / Format: CCP4 / Size: 166.4 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_22615.map.gz / Format: CCP4 / Size: 166.4 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Post-processed full map | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.36 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
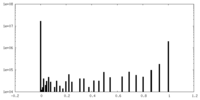
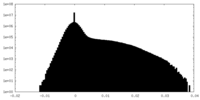
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Mask #1
| File |  emd_22615_msk_1.map emd_22615_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
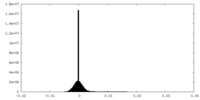
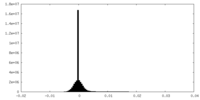
| Density Histograms |
-Additional map: Unfiltered and unmasked half map1 up to Nyquist frequency
| File | emd_22615_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Unfiltered and unmasked half map1 up to Nyquist frequency | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: Unfiltered and unmasked half map2 up to Nyquist frequency
| File | emd_22615_additional_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Unfiltered and unmasked half map2 up to Nyquist frequency | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Masked half map1
| File | emd_22615_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Masked half map1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Masked half map2
| File | emd_22615_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Masked half map2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Rabbit RyR1 with Mg2+ and AMP-PCP
| Entire | Name: Rabbit RyR1 with Mg2+ and AMP-PCP |
|---|---|
| Components |
|
-Supramolecule #1: Rabbit RyR1 with Mg2+ and AMP-PCP
| Supramolecule | Name: Rabbit RyR1 with Mg2+ and AMP-PCP / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 Details: Purified RyR1 was reconstituted with membrane scaffold protein MSP1E3D1, and POPC. |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 2.26 MDa |
-Macromolecule #1: RyR1
| Macromolecule | Name: RyR1 / type: protein_or_peptide / ID: 1 / Number of copies: 4 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 533.66325 KDa |
| Sequence | String: MGDGGEGEDE VQFLRTDDEV VLQCSATVLK EQLKLCLAAE GFGNRLCFLE PTSNAQNVPP DLAICCFTLE QSLSVRALQE MLANTVEAG VESSQGGGHR TLLYGHAILL RHAHSRMYLS CLTTSRSMTD KLAFDVGLQE DATGEACWWT MHPASKQRSE G EKVRVGDD ...String: MGDGGEGEDE VQFLRTDDEV VLQCSATVLK EQLKLCLAAE GFGNRLCFLE PTSNAQNVPP DLAICCFTLE QSLSVRALQE MLANTVEAG VESSQGGGHR TLLYGHAILL RHAHSRMYLS CLTTSRSMTD KLAFDVGLQE DATGEACWWT MHPASKQRSE G EKVRVGDD LILVSVSSER YLHLSTASGE LQVDASFMQT LWNMNPICSC CEEGYVTGGH VLRLFHGHMD ECLTISAADS DD QRRLVYY EGGAVCTHAR SLWRLEPLRI SWSGSHLRWG QPLRIRHVTT GRYLALTEDQ GLVVVDACKA HTKATSFCFR VSK EKLDTA PKRDVEGMGP PEIKYGESLC FVQHVASGLW LTYAAPDPKA LRLGVLKKKA ILHQEGHMDD ALFLTRCQQE ESQA ARMIH STAGLYNQFI KGLDSFSGKP RGSGPPAGPA LPIEAVILSL QDLIGYFEPP SEELQHEEKQ SKLRSLRNRQ SLFQE EGML SLVLNCIDRL NVYTTAAHFA EYAGEEAAES WKEIVNLLYE LLASLIRGNR ANCALFSTNL DWVVSKLDRL EASSGI LEV LYCVLIESPE VLNIIQENHI KSIISLLDKH GRNHKVLDVL CSLCVCNGVA VRSNQDLITE NLLPGRELLL QTNLINY VT SIRPNIFVGR AEGSTQYGKW YFEVMVDEVV PFLTAQATHL RVGWALTEGY SPYPGGGEGW GGNGVGDDLY SYGFDGLH L WTGHVARPVT SPGQHLLAPE DVVSCCLDLS VPSISFRING CPVQGVFEAF NLDGLFFPVV SFSAGVKVRF LLGGRHGEF KFLPPPGYAP CHEAVLPRER LRLEPIKEYR REGPRGPHLV GPSRCLSHTD FVPCPVDTVQ IVLPPHLERI REKLAENIHE LWALTRIEQ GWTYGPVRDD NKRLHPCLVN FHSLPEPERN YNLQMSGETL KTLLALGCHV GMADEKAEDN LKKTKLPKTY M MSNGYKPA PLDLSHVRLT PAQTTLVDRL AENGHNVWAR DRVAQGWSYS AVQDIPARRN PRLVPYRLLD EATKRSNRDS LC QAVRTLL GYGYNIEPPD QEPSQVENQS RWDRVRIFRA EKSYTVQSGR WYFEFEAVTT GEMRVGWARP ELRPDVELGA DEL AYVFNG HRGQRWHLGS EPFGRPWQSG DVVGCMIDLT ENTIIFTLNG EVLMSDSGSE TAFREIEIGD GFLPVCSLGP GQVG HLNLG QDVSSLRFFA ICGLQEGFEP FAINMQRPVT TWFSKSLPQF EPVPPEHPHY EVARMDGTVD TPPCLRLAHR (UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) EL GKQKNIMPLS AAMFLSERKN PAPQCPPRLE VQMLMPVSWS RMPNHFLQVE TRRAGERLGW AVQCQDPLTM MALHIPEE N RCMDILELSE RLDLQRFHSH TLRLYRAVCA LGNNRVAHAL CSHVDQAQLL HALEDAHLPG PLRAGYYDLL ISIHLESAC RSRRSMLSEY IVPLTPETRA ITLFPPGRKG GNARRHGLPG VGVTTSLRPP HHFSPPCFVA ALPAAGVAEA PARLSPAIPL EALRDKALR MLGEAVRDGG QHARDPVGGS VEFQFVPVLK LVSTLLVMGI FGDEDVKQIL KMIEPEVFTE EEEEEEEEEE E EEEEEEDE EEKEEDEEEE EKEDAEKEEE EAPEGEKEDL EEGLLQMKLP ESVKLQMCNL LEYFCDQELQ HRVESLAAFA ER YVDKLQA NQRSRYALLM RAFTMSAAET ARRTREFRSP PQEQINMLLH FKDEADEEDC PLPEDIRQDL QDFHQDLLAH CGI QLEGEE EEPEEETSLS SRLRSLLETV RLVKKKEEKP EEELPAEEKK PQSLQELVSH MVVRWAQEDY VQSPELVRAM FSLL HRQYD GLGELLRALP RAYTISPSSV EDTMSLLECL GQIRSLLIVQ MGPQEENLMI QSIGNIMNNK VFYQHPNLMR ALGMH ETVM EVMVNVLGGG ETKEIRFPKM VTSCCRFLCY FCRISRQNQR SMFDHLSYLL ENSGIGLGMQ GSTPLDVAAA SVIDNN ELA LALQEQDLEK VVSYLAGCGL QSCPMLLAKG YPDIGWNPCG GERYLDFLRF AVFVNGESVE ENANVVVRLL IRKPECF GP ALRGEGGSGL LAAIEEAIRI SEDPARDGPG VRRDRRREHF GEEPPEENRV HLGHAIMSFY AALIDLLGRC APEMHLIQ A GKGEALRIRA ILRSLVPLDD LVGIISLPLQ IPTLGKDGAL V(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)CAIAGALP PDYVDASYSS KAEKKATVDA EGNFDPRPVE TLNVIIPEKL DSFINKFAEY THEKWAFDKI QN NWSYGEN VDEELKTHPM LRPYKTFSEK DKEIYRWPIK ESLKAMIAWE WTIEKAREGE EERTEKKKTR KISQTAQTYD PRE GYNPQP PDLSGVTLSR ELQAMAEQLA ENYHNTWGRK KKQELEAKGG GTHPLLVPYD TLTAKEKARD REKAQELLKF LQMN GYAVT RGL(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK) (UNK)(UNK)KSK KAVWHKLLSK QRRRAVVACF RMTPLYNLPT HRACNMFLES YKAAWILTED HSFEDRMIDD LSKAGEQ EE EEEEVEEKKP DPLHQLVLHF SRTALTEKSK LDEDYLYMAY ADIMAKSCHL EEGGENGEAE EEEVEVSFEE KEMEKQRL L YQQSRLHTRG AAEMVLQMIS ACKGETGAMV SSTLKLGISI LNGGNAEVQQ KMLDYLKDKK EVGFFQSIQA LMQTCSVLD LNAFERQNKA EGLGMVNEDG TVINRQNGEK VMADDEFTQD LFRFLQLLCE GHNNDFQNYL RTQTGNTTTI NIIICTVDYL LRLQESISD FYWYYSGKDV IEEQGKRNFS KAMSVAKQVF NSLTEYIQGP CTGNQQSLAH SRLWDAVVGF LHVFAHMMMK L AQDSSQIE LLKELLDLQK DMVVMLLSLL EGNVVNGMIA RQMVDMLVES SSNVEMILKF FDMFLKLKDI VGSEAFQDYV TD PRGLISK KDFQKAMDSQ KQFTGPEIQF LLSCSEADEN EMINFEEFAN RFQEPARDIG FNVAVLLTNL SEHVPHDPRL RNF LELAES ILEYFRPYLG RIEIMGASRR IERIYFEISE TNRAQWEMPQ VKESKRQFIF DVVNEGGEAE KMELFVSFCE DTIF EMQIA AQISEPEGEP EADEDEGMGE AAAEGAEEGA AGAEGAAGTV AAGATARLAA AAARALRGLS YRSLRRRVRR LRRLT AREA ATALAALLWA VVARAGAAGA GAAAGALRLL WGSLFGGGLV EGAKKVTVTE LLAGMPDPTS DEVHGEQPAG PGGDAD GAG EGEGEGDAAE GDGDEEVAGH EAGPGGAEGV VAVADGGPFR PEGAGGLGDM GDTTPAEPPT PEGSPILKRK LGVDGEE EE LVPEPEPEPE PEPEKADEEN GEKEEVPEAP PEPPKKAPPS PPAKKEEAGG AGMEFWGELE VQRVKFLNYL SRNFYTLR F LALFLAFAIN FILLFYKVSD SPPGEDDMEG SAAGDLAGAG SGGGSGWGSG AGEEAEGDED ENMVYYFLEE STGYMEPAL WCLSLLHTLV AFLCIIGYNC LKVPLVIFKR EKELARKLEF DGLYITEQPG DDDVKGQWDR LVLNTPSFPS NYWDKFVKRK VLDKHGDIF GRERIAELLG MDLASLEITA HNERKPDPPP GLLTWLMSID VKYQIWKFGV IFTDNSFLYL GWYMVMSLLG H YNNFFFAA HLLDIAMGVK TLRTILSSVT HNGKQLVMTV GLLAVVVYLY TVVAFNFFRK FYNKSEDEDE PDMKCDDMMT CY LFHMYVG VRAGGGIGDE IEDPAGDEYE LYRVVFDITF FFFVIVILLA IIQGLIIDAF GELRDQQEQV KEDMETKCFI CGI GSDYFD TTPHGFETHT LEEHNLANYM FFLMYLINKD ETEHTGQESY VWKMYQERCW DFFPAGDCFR KQYEDQLS |
-Macromolecule #2: PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER
| Macromolecule | Name: PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER / type: ligand / ID: 2 / Number of copies: 4 / Formula: ACP |
|---|---|
| Molecular weight | Theoretical: 505.208 Da |
| Chemical component information |  ChemComp-ACP: |
-Macromolecule #3: ZINC ION
| Macromolecule | Name: ZINC ION / type: ligand / ID: 3 / Number of copies: 4 / Formula: ZN |
|---|---|
| Molecular weight | Theoretical: 65.409 Da |
-Macromolecule #4: MAGNESIUM ION
| Macromolecule | Name: MAGNESIUM ION / type: ligand / ID: 4 / Number of copies: 5 / Formula: MG |
|---|---|
| Molecular weight | Theoretical: 24.305 Da |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 4.2 mg/mL | |||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.4 Component:
| |||||||||||||||||||||
| Grid | Model: C-flat-2/1 / Material: GOLD / Mesh: 300 / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 30 sec. / Pretreatment - Atmosphere: AIR | |||||||||||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 95 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV Details: Sample was blotted for 1 second on both sides with Whatman hardened ashless filter paper with blot force 2.. | |||||||||||||||||||||
| Details | Purified RyR1 was reconstituted with membrane scaffold protein MSP1E3D1, and POPC in 1:2:50 molar ratio. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Specialist optics | Energy filter - Slit width: 20 eV |
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: SUPER-RESOLUTION / Number grids imaged: 2 / Number real images: 3371 / Average electron dose: 60.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 100.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.5 µm / Nominal defocus min: 1.0 µm / Nominal magnification: 105000 |
| Sample stage | Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: FLEXIBLE FIT |
|---|---|
| Output model |  PDB-7k0s: |
 Movie
Movie Controller
Controller

















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)