[English] 日本語
 Yorodumi
Yorodumi- EMDB-26610: Cryo-EM structure of rabbit RyR1 in the presence of high Mg2+ and... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of rabbit RyR1 in the presence of high Mg2+ and AMP-PCP in nanodisc | |||||||||||||||||||||
 Map data Map data | ||||||||||||||||||||||
 Sample Sample |
| |||||||||||||||||||||
 Keywords Keywords | Ryanodine Receptor / RyR1 / Intracellular Calcium channel / Mg2+ / Inhibition / Excitation-Contraction coupling / TRANSPORT PROTEIN | |||||||||||||||||||||
| Biological species |  | |||||||||||||||||||||
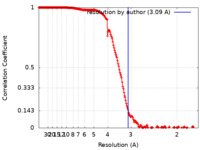
| Method | single particle reconstruction / cryo EM / Resolution: 3.09 Å | |||||||||||||||||||||
 Authors Authors | Nayak AR / Samso M | |||||||||||||||||||||
| Funding support |  United States, 6 items United States, 6 items
| |||||||||||||||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2024 Journal: Nat Commun / Year: 2024Title: Interplay between Mg2+ and Ca2+ at multiple sites of the ryanodine receptor Authors: Nayak AR / Rangubpit W / Will AH / Hu Y / Castro-Hartmann P / Lobo JJ / Dryden K / Lamb GD / Sompornpisut P / Samso M | |||||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_26610.map.gz emd_26610.map.gz | 500.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-26610-v30.xml emd-26610-v30.xml emd-26610.xml emd-26610.xml | 26.7 KB 26.7 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_26610_fsc.xml emd_26610_fsc.xml | 19.2 KB | Display |  FSC data file FSC data file |
| Images |  emd_26610.png emd_26610.png | 132.9 KB | ||
| Filedesc metadata |  emd-26610.cif.gz emd-26610.cif.gz | 9.2 KB | ||
| Others |  emd_26610_half_map_1.map.gz emd_26610_half_map_1.map.gz emd_26610_half_map_2.map.gz emd_26610_half_map_2.map.gz | 491.7 MB 491.7 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-26610 http://ftp.pdbj.org/pub/emdb/structures/EMD-26610 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-26610 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-26610 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_26610.map.gz / Format: CCP4 / Size: 641.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_26610.map.gz / Format: CCP4 / Size: 641.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.86 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_26610_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
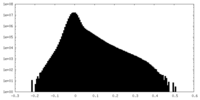
| Projections & Slices |
| ||||||||||||
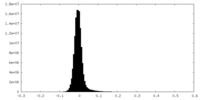
| Density Histograms |
-Half map: #1
| File | emd_26610_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
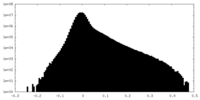
| Projections & Slices |
| ||||||||||||
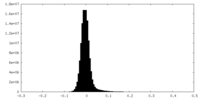
| Density Histograms |
- Sample components
Sample components
-Entire : Rabbit RyR1 with AMP-PCP and inhibiting Mg2+
| Entire | Name: Rabbit RyR1 with AMP-PCP and inhibiting Mg2+ |
|---|---|
| Components |
|
-Supramolecule #1: Rabbit RyR1 with AMP-PCP and inhibiting Mg2+
| Supramolecule | Name: Rabbit RyR1 with AMP-PCP and inhibiting Mg2+ / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 Details: Purified RyR1 was reconstituted with membrane scaffold protein, MSP1E3D1 and POPC. |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 2.26 MDa |
-Macromolecule #1: Ryanodine receptor 1
| Macromolecule | Name: Ryanodine receptor 1 / type: protein_or_peptide / ID: 1 / Number of copies: 4 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 542.887 KDa |
| Sequence | String: MGDGGEGEDE VQFLRTDDEV VLQCSATVLK EQLKLCLAAE GFGNRLCFLE PTSNAQNVPP DLAICCFTLE QSLSVRALQE MLANTVEAG VESSQGGGHR TLLYGHAILL RHAHSRMYLS CLTTSRSMTD KLAFDVGLQE DATGEACWWT MHPASKQRSE G EKVRVGDD ...String: MGDGGEGEDE VQFLRTDDEV VLQCSATVLK EQLKLCLAAE GFGNRLCFLE PTSNAQNVPP DLAICCFTLE QSLSVRALQE MLANTVEAG VESSQGGGHR TLLYGHAILL RHAHSRMYLS CLTTSRSMTD KLAFDVGLQE DATGEACWWT MHPASKQRSE G EKVRVGDD LILVSVSSER YLHLSTASGE LQVDASFMQT LWNMNPICSC CEEGYVTGGH VLRLFHGHMD ECLTISAADS DD QRRLVYY EGGAVCTHAR SLWRLEPLRI SWSGSHLRWG QPLRIRHVTT GRYLALTEDQ GLVVVDACKA HTKATSFCFR VSK EKLDTA PKRDVEGMGP PEIKYGESLC FVQHVASGLW LTYAAPDPKA LRLGVLKKKA ILHQEGHMDD ALFLTRCQQE ESQA ARMIH STAGLYNQFI KGLDSFSGKP RGSGPPAGPA LPIEAVILSL QDLIGYFEPP SEELQHEEKQ SKLRSLRNRQ SLFQE EGML SLVLNCIDRL NVYTTAAHFA EYAGEEAAES WKEIVNLLYE LLASLIRGNR ANCALFSTNL DWVVSKLDRL EASSGI LEV LYCVLIESPE VLNIIQENHI KSIISLLDKH GRNHKVLDVL CSLCVCNGVA VRSNQDLITE NLLPGRELLL QTNLINY VT SIRPNIFVGR AEGSTQYGKW YFEVMVDEVV PFLTAQATHL RVGWALTEGY SPYPGGGEGW GGNGVGDDLY SYGFDGLH L WTGHVARPVT SPGQHLLAPE DVVSCCLDLS VPSISFRING CPVQGVFEAF NLDGLFFPVV SFSAGVKVRF LLGGRHGEF KFLPPPGYAP CHEAVLPRER LRLEPIKEYR REGPRGPHLV GPSRCLSHTD FVPCPVDTVQ IVLPPHLERI REKLAENIHE LWALTRIEQ GWTYGPVRDD NKRLHPCLVN FHSLPEPERN YNLQMSGETL KTLLALGCHV GMADEKAEDN LKKTKLPKTY M MSNGYKPA PLDLSHVRLT PAQTTLVDRL AENGHNVWAR DRVAQGWSYS AVQDIPARRN PRLVPYRLLD EATKRSNRDS LC QAVRTLL GYGYNIEPPD QEPSQVENQS RWDRVRIFRA EKSYTVQSGR WYFEFEAVTT GEMRVGWARP ELRPDVELGA DEL AYVFNG HRGQRWHLGS EPFGRPWQSG DVVGCMIDLT ENTIIFTLNG EVLMSDSGSE TAFREIEIGD GFLPVCSLGP GQVG HLNLG QDVSSLRFFA ICGLQEGFEP FAINMQRPVT TWFSKSLPQF EPVPPEHPHY EVARMDGTVD TPPCLRLAHR TWGSQ NSLV EMLFLRLSLP VQFHQHFRCT AGATPLAPPG LQPPAEDEAR AAEPDPDYEN LRRSAGGWGE AEGGKEGTAK EGTPGG TPQ PGVEAQPVRA ENEKDATTEK NKKRGFLFKA KKAAMMTQPP ATPALPRLPH DVVPADNRDD PEIILNTTTY YYSVRVF AG QEPSCVWVGW VTPDYHQHDM NFDLSKVRAV TVTMGDEQGN VHSSLKC(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)A(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK) (UNK)(UNK)(UNK)(UNK)(UNK)ELG KQKNIMPLSA AMFLSERKNP APQCPPRLEV QMLMPVSWSR MPNHFLQVET RR AGERLGW AVQCQDPLTM MALHIPEENR CMDILELSER LDLQRFHSHT LRLYRAVCAL GNNRVAHALC SHVDQAQLLH ALE DAHLPG PLRAGYYDLL ISIHLESACR SRRSMLSEYI VPLTPETRAI TLFPPGRKGG NARRHGLPGV GVTTSLRPPH HFSP PCFVA ALPAAGVAEA PARLSPAIPL EALRDKALRM LGEAVRDGGQ HARDPVGGSV EFQFVPVLKL VSTLLVMGIF GDEDV KQIL KMIEPEVFTE EEEEEEEEEE EEEEEEEDEE EKEEDEEEEE KEDAEKEEEE APEGEKEDLE EGLLQMKLPE SVKLQM CNL LEYFCDQELQ HRVESLAAFA ERYVDKLQAN QRSRYALLMR AFTMSAAETA RRTREFRSPP QEQINMLLHF KDEADEE DC PLPEDIRQDL QDFHQDLLAH CGIQLEGEEE EPEEETSLSS RLRSLLETVR LVKKKEEKPE EELPAEEKKP QSLQELVS H MVVRWAQEDY VQSPELVRAM FSLLHRQYDG LGELLRALPR AYTISPSSVE DTMSLLECLG QIRSLLIVQM GPQEENLMI QSIGNIMNNK VFYQHPNLMR ALGMHETVME VMVNVLGGGE TKEIRFPKMV TSCCRFLCYF CRISRQNQRS MFDHLSYLLE NSGIGLGMQ GSTPLDVAAA SVIDNNELAL ALQEQDLEKV VSYLAGCGLQ SCPMLLAKGY PDIGWNPCGG ERYLDFLRFA V FVNGESVE ENANVVVRLL IRKPECFGPA LRGEGGSGLL AAIEEAIRIS EDPARDGPGV RRDRRREHFG EEPPEENRVH LG HAIMSFY AALIDLLGRC APEMHLIQAG KGEALRIRAI LRSLVPLDDL VGIISLPLQI PTLGKDGALV Q(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK) (UNK)(UNK)MPC LCAIAGALPP DYVDASYSSK AEKKATVDAE GNFDPRPVET LNVIIPEKLD SFINKFAEYT HEKWAFD KI QNNWSYGENV DEELKTHPML RPYKTFSEKD KEIYRWPIKE SLKAMIAWEW TIEKAREGEE ERTEKKKTRK ISQTAQTY D PREGYNPQPP DLSGVTLSRE LQAMAEQLAE NYHNTWGRKK KQELEAKGGG THPLLVPYDT LTAKEKARDR EKAQELLKF LQMNGYAVTR GL(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)L EAVVSSGRVE KSPHEQE(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) VVNCL HILA RSLDARTVMK SGPEIVKAGL RSFFESASED IEKMVENLRL GKVSQARTQV KGVGQNLTYT T(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)SCYRTL(UNK) (UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) EEMCPDIPV LDRLMADIGG LAESG(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)FLTA DSKSKMAKAG DAQSGGSDQE RTKKKRRGDR YSVQTSLI(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)KS KKAVWHKLLS KQRRRAVVAC FRMTPLYNLP THRACNMFLE SYKAAWILTE DHS FEDRMI DDLSKAGEQE EEEEEVEEKK PDPLHQLVLH FSRTALTEKS KLDEDYLYMA YADIMAKSCH LEEGGENGEA EEEE VEVSF EEKEMEKQRL LYQQSRLHTR GAAEMVLQMI SACKGETGAM VSSTLKLGIS ILNGGNAEVQ QKMLDYLKDK KEVGF FQSI QALMQTCSVL DLNAFERQNK AEGLGMVNED GTVINRQNGE KVMADDEFTQ DLFRFLQLLC EGHNNDFQNY LRTQTG NTT TINIIICTVD YLLRLQESIS DFYWYYSGKD VIEEQGKRNF SKAMSVAKQV FNSLTEYIQG PCTGNQQSLA HSRLWDA VV GFLHVFAHMM MKLAQDSSQI ELLKELLDLQ KDMVVMLLSL LEGNVVNGMI ARQMVDMLVE SSSNVEMILK FFDMFLKL K DIVGSEAFQD YVTDPRGLIS KKDFQKAMDS QKQFTGPEIQ FLLSCSEADE NEMINFEEFA NRFQEPARDI GFNVAVLLT NLSEHVPHDP RLRNFLELAE SILEYFRPYL GRIEIMGASR RIERIYFEIS ETNRAQWEMP QVKESKRQFI FDVVNEGGEA EKMELFVSF CEDTIFEMQI AAQISEPEGE PEADEDEGMG EAAAEGAEEG AAGAEGAAGT VAAGATARLA AAAARALRGL S YRSLRRRV RRLRRLTARE AATALAALLW AVVARAGAAG AGAAAGALRL LWGSLFGGGL VEGAKKVTVT ELLAGMPDPT SD EVHGEQP AGPGGDADGA GEGEGEGDAA EGDGDEEVAG HEAGPGGAEG VVAVADGGPF RPEGAGGLGD MGDTTPAEPP TPE GSPILK RKLGVDGEEE ELVPEPEPEP EPEPEKADEE NGEKEEVPEA PPEPPKKAPP SPPAKKEEAG GAGMEFWGEL EVQR VKFLN YLSRNFYTLR FLALFLAFAI NFILLFYKVS DSPPGEDDME GSAAGDLAGA GSGGGSGWGS GAGEEAEGDE DENMV YYFL EESTGYMEPA LWCLSLLHTL VAFLCIIGYN CLKVPLVIFK REKELARKLE FDGLYITEQP GDDDVKGQWD RLVLNT PSF PSNYWDKFVK RKVLDKHGDI FGRERIAELL GMDLASLEIT AHNERKPDPP PGLLTWLMSI DVKYQIWKFG VIFTDNS FL YLGWYMVMSL LGHYNNFFFA AHLLDIAMGV KTLRTILSSV THNGKQLVMT VGLLAVVVYL YTVVAFNFFR KFYNKSED E DEPDMKCDDM MTCYLFHMYV GVRAGGGIGD EIEDPAGDEY ELYRVVFDIT FFFFVIVILL AIIQGLIIDA FGELRDQQE QVKEDMETKC FICGIGSDYF DTTPHGFETH TLEEHNLANY MFFLMYLINK DETEHTGQES YVWKMYQERC WDFFPAGDCF RKQYEDQLS |
-Macromolecule #2: PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER
| Macromolecule | Name: PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER / type: ligand / ID: 2 / Number of copies: 4 / Formula: ACP |
|---|---|
| Molecular weight | Theoretical: 505.208 Da |
| Chemical component information |  ChemComp-ACP: |
-Macromolecule #3: ZINC ION
| Macromolecule | Name: ZINC ION / type: ligand / ID: 3 / Number of copies: 4 / Formula: ZN |
|---|---|
| Molecular weight | Theoretical: 65.409 Da |
-Macromolecule #4: MAGNESIUM ION
| Macromolecule | Name: MAGNESIUM ION / type: ligand / ID: 4 / Number of copies: 13 / Formula: MG |
|---|---|
| Molecular weight | Theoretical: 24.305 Da |
-Macromolecule #5: 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine
| Macromolecule | Name: 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine / type: ligand / ID: 5 / Number of copies: 8 / Formula: LBN |
|---|---|
| Molecular weight | Theoretical: 760.076 Da |
| Chemical component information |  ChemComp-LBN: |
-Macromolecule #6: water
| Macromolecule | Name: water / type: ligand / ID: 6 / Number of copies: 8 / Formula: HOH |
|---|---|
| Molecular weight | Theoretical: 18.015 Da |
| Chemical component information |  ChemComp-HOH: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 4.2 mg/mL |
|---|---|
| Buffer | pH: 7.4 Details: 20 mM MOPS (pH=7.4), 635mM KCl, 2mM DTT, 11.6mM MgCl2, 1mM AMP-PCP |
| Grid | Model: UltrAuFoil R1.2/1.3 / Material: GOLD / Mesh: 300 / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 30 sec. / Pretreatment - Atmosphere: AIR |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV |
| Details | Purified RyR1 was reconstituted with membrane scaffold protein MSP1E3D1 and POPC at a 1:2:50 molar ratio. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Specialist optics | Energy filter - Slit width: 10 eV |
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Number real images: 9140 / Average electron dose: 60.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 100.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.0 µm / Nominal defocus min: 1.0 µm / Nominal magnification: 105000 |
| Sample stage | Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: FLEXIBLE FIT |
|---|---|
| Output model |  PDB-7umz: |
 Movie
Movie Controller
Controller





 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)