[English] 日本語
 Yorodumi
Yorodumi- EMDB-12682: Cryo-EM structure (model_2b) of the RC-dLH complex from Gemmatimo... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-12682 | ||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure (model_2b) of the RC-dLH complex from Gemmatimonas phototrophica at 2.44 A | ||||||||||||||||||||||||
 Map data Map data | |||||||||||||||||||||||||
 Sample Sample |
| ||||||||||||||||||||||||
 Keywords Keywords | Photosynthesis / reaction centre light harvesting complex / RC-dLH / RC-LH1 / carotenoid / quinone / bacteriochlorophyll a / Gemmatimonas phototrophica / MEMBRANE PROTEIN | ||||||||||||||||||||||||
| Function / homology |  Function and homology information Function and homology informationorganelle inner membrane / plasma membrane light-harvesting complex / bacteriochlorophyll binding / photosynthetic electron transport in photosystem II / photosynthesis, light reaction / endomembrane system / electron transfer activity / iron ion binding / heme binding / metal ion binding ...organelle inner membrane / plasma membrane light-harvesting complex / bacteriochlorophyll binding / photosynthetic electron transport in photosystem II / photosynthesis, light reaction / endomembrane system / electron transfer activity / iron ion binding / heme binding / metal ion binding / membrane / plasma membrane Similarity search - Function | ||||||||||||||||||||||||
| Biological species |  Gemmatimonas phototrophica (bacteria) Gemmatimonas phototrophica (bacteria) | ||||||||||||||||||||||||
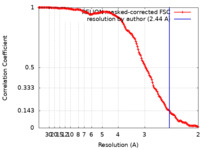
| Method | single particle reconstruction / cryo EM / Resolution: 2.44 Å | ||||||||||||||||||||||||
 Authors Authors | Qian P / Koblizek M | ||||||||||||||||||||||||
| Funding support |  Czech Republic, Czech Republic,  United Kingdom, European Union, 7 items United Kingdom, European Union, 7 items
| ||||||||||||||||||||||||
 Citation Citation |  Journal: Sci Adv / Year: 2022 Journal: Sci Adv / Year: 2022Title: 2.4-Å structure of the double-ring photosystem. Authors: Pu Qian / Alastair T Gardiner / Ivana Šímová / Katerina Naydenova / Tristan I Croll / Philip J Jackson / Nupur / Miroslav Kloz / Petra Čubáková / Marek Kuzma / Yonghui Zeng / Pablo ...Authors: Pu Qian / Alastair T Gardiner / Ivana Šímová / Katerina Naydenova / Tristan I Croll / Philip J Jackson / Nupur / Miroslav Kloz / Petra Čubáková / Marek Kuzma / Yonghui Zeng / Pablo Castro-Hartmann / Bart van Knippenberg / Kenneth N Goldie / David Kaftan / Pavel Hrouzek / Jan Hájek / Jon Agirre / C Alistair Siebert / David Bína / Kasim Sader / Henning Stahlberg / Roman Sobotka / Christopher J Russo / Tomáš Polívka / C Neil Hunter / Michal Koblížek /     Abstract: Phototrophic Gemmatimonadetes evolved the ability to use solar energy following horizontal transfer of photosynthesis-related genes from an ancient phototrophic proteobacterium. The electron cryo- ...Phototrophic Gemmatimonadetes evolved the ability to use solar energy following horizontal transfer of photosynthesis-related genes from an ancient phototrophic proteobacterium. The electron cryo-microscopy structure of the photosystem at 2.4 Å reveals a unique, double-ring complex. Two unique membrane-extrinsic polypeptides, RC-S and RC-U, hold the central type 2 reaction center (RC) within an inner 16-subunit light-harvesting 1 (LH1) ring, which is encircled by an outer 24-subunit antenna ring (LHh) that adds light-gathering capacity. Femtosecond kinetics reveal the flow of energy within the RC-dLH complex, from the outer LHh ring to LH1 and then to the RC. This structural and functional study shows that has independently evolved its own compact, robust, and highly effective architecture for harvesting and trapping solar energy. | ||||||||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_12682.map.gz emd_12682.map.gz | 19.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-12682-v30.xml emd-12682-v30.xml emd-12682.xml emd-12682.xml | 33.6 KB 33.6 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_12682_fsc.xml emd_12682_fsc.xml | 14.1 KB | Display |  FSC data file FSC data file |
| Images |  emd_12682.png emd_12682.png | 95.9 KB | ||
| Filedesc metadata |  emd-12682.cif.gz emd-12682.cif.gz | 9.6 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-12682 http://ftp.pdbj.org/pub/emdb/structures/EMD-12682 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-12682 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-12682 | HTTPS FTP |
-Related structure data
| Related structure data |  7o0xMC  7o0uC  7o0vC  7o0wC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | |
| EM raw data |  EMPIAR-10681 (Title: Movies of a photosynthetic reaction centre - double light-harvesting 1 complex from the bacterium G. phototrophica EMPIAR-10681 (Title: Movies of a photosynthetic reaction centre - double light-harvesting 1 complex from the bacterium G. phototrophicaData size: 12.1 TB Data #1: Unaligned multi-frame micrographs of RC-dLH1 complexes on HexAuFoil support [micrographs - multiframe]) |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_12682.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_12682.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.99946 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
+Entire : RC-dLH model_2b
+Supramolecule #1: RC-dLH model_2b
+Macromolecule #1: LHh-alpha
+Macromolecule #2: Light-harvesting protein B:885 subunit beta
+Macromolecule #3: MULTIHEME_CYTC domain-containing protein
+Macromolecule #4: RC-S
+Macromolecule #5: RC-U
+Macromolecule #6: PRCH domain-containing protein
+Macromolecule #7: RC-Hc
+Macromolecule #8: Photosynthetic reaction center L subunit
+Macromolecule #9: RC-M
+Macromolecule #10: LHC domain-containing protein
+Macromolecule #11: LHC domain-containing protein
+Macromolecule #13: BACTERIOCHLOROPHYLL A
+Macromolecule #14: DODECYL-BETA-D-MALTOSIDE
+Macromolecule #15: (2~{E},4~{E},6~{E},10~{E},12~{E},14~{E},16~{E},18~{E},20~{E},22~{...
+Macromolecule #16: HEME C
+Macromolecule #17: 2-acetamido-2-deoxy-alpha-D-glucopyranose
+Macromolecule #18: (2~{S},3~{S},4~{S},5~{S})-4,5-diacetyloxy-3-oxidanyl-oxane-2-carb...
+Macromolecule #19: (1R)-2-{[(S)-{[(2S)-2,3-dihydroxypropyl]oxy}(hydroxy)phosphoryl]o...
+Macromolecule #20: (19R,22S)-25-amino-22-hydroxy-22-oxido-16-oxo-17,21,23-trioxa-22l...
+Macromolecule #21: (2R,5R,11R,14R)-5,8,11-trihydroxy-5,11-dioxido-17-oxo-2,14-bis(te...
+Macromolecule #22: BACTERIOPHEOPHYTIN A
+Macromolecule #23: MENAQUINONE 8
+Macromolecule #24: FE (III) ION
+Macromolecule #25: SPIRILLOXANTHIN
+Macromolecule #26: [(2~{S})-3-[(2~{R},3~{R},4~{R},5~{S},6~{R})-6-(hydroxymethyl)-5-[...
+Macromolecule #27: [(2~{S})-3-[(2~{R},3~{R},4~{S},5~{S},6~{R})-6-(hydroxymethyl)-3,4...
+Macromolecule #28: water
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 6.5 mg/mL | ||||||
|---|---|---|---|---|---|---|---|
| Buffer | pH: 8 / Component:
Details: The final purified complex is in 20mM Tris.Cl, 0.025 DDM, PH 8.0 buffer. | ||||||
| Grid | Model: Homemade / Material: GOLD / Support film - Material: GOLD / Support film - topology: HOLEY ARRAY / Support film - Film thickness: 30 / Pretreatment - Type: PLASMA CLEANING / Pretreatment - Time: 120 sec. / Pretreatment - Atmosphere: OTHER | ||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: HOMEMADE PLUNGER / Details: Talmon type. | ||||||
| Details | The protein complex was isolated from photosynthetic membrane using detergent beta-DDM. After purification, concentrated protein sample solution was stored in LN before using. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON IV (4k x 4k) / Number grids imaged: 1 / Number real images: 19865 / Average exposure time: 1.2 sec. / Average electron dose: 24.8 e/Å2 Details: All movies were recorded from a HexAuFoil grid with a special EPU version, which can recognise 300 nm holes on the grid. |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: -2.4 µm / Nominal defocus min: -0.8 µm / Nominal magnification: 120000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Initial model |
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Details | ISOLDE incorporated in ChimeraX was used. | ||||||||
| Refinement | Space: REAL / Protocol: FLEXIBLE FIT / Overall B value: 30 | ||||||||
| Output model |  PDB-7o0x: |
 Movie
Movie Controller
Controller














 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)