[English] 日本語
 Yorodumi
Yorodumi- EMDB-15863: Cryo-EM structure of ribosome-Sec61 in complex with cyclotriazadi... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of ribosome-Sec61 in complex with cyclotriazadisulfonamide derivative CK147 | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
| Function / homology |  Function and homology information Function and homology informationSsh1 translocon complex / ribosomal subunit / post-translational protein targeting to membrane, translocation / protein transmembrane transporter activity / ubiquitin ligase inhibitor activity / positive regulation of signal transduction by p53 class mediator / protein-RNA complex assembly / rough endoplasmic reticulum / MDM2/MDM4 family protein binding / cytosolic ribosome ...Ssh1 translocon complex / ribosomal subunit / post-translational protein targeting to membrane, translocation / protein transmembrane transporter activity / ubiquitin ligase inhibitor activity / positive regulation of signal transduction by p53 class mediator / protein-RNA complex assembly / rough endoplasmic reticulum / MDM2/MDM4 family protein binding / cytosolic ribosome / ribosomal large subunit biogenesis / maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA) / antimicrobial humoral immune response mediated by antimicrobial peptide / regulation of translation / large ribosomal subunit / ribosome binding / ribosomal large subunit assembly / 5S rRNA binding / large ribosomal subunit rRNA binding / cytosolic large ribosomal subunit / killing of cells of another organism / defense response to Gram-negative bacterium / cytoplasmic translation / tRNA binding / rRNA binding / postsynaptic density / structural constituent of ribosome / ribosome / translation / ribonucleoprotein complex / mRNA binding / synapse / nucleolus / endoplasmic reticulum / RNA binding / zinc ion binding / nucleus / cytoplasm / cytosol Similarity search - Function | |||||||||
| Biological species |     | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 2.79 Å | |||||||||
 Authors Authors | Pauwels E / Shewakramani NR / De Wijngaert B / Vermeire K / Das K | |||||||||
| Funding support |  Belgium, 1 items Belgium, 1 items
| |||||||||
 Citation Citation |  Journal: Sci Adv / Year: 2023 Journal: Sci Adv / Year: 2023Title: Structural insights into TRAP association with ribosome-Sec61 complex and translocon inhibition by a CADA derivative. Authors: Eva Pauwels / Neesha R Shewakramani / Brent De Wijngaert / Anita Camps / Becky Provinciael / Joren Stroobants / Kai-Uwe Kalies / Enno Hartmann / Piet Maes / Kurt Vermeire / Kalyan Das /   Abstract: During cotranslational translocation, the signal peptide of a nascent chain binds Sec61 translocon to initiate protein transport through the endoplasmic reticulum (ER) membrane. Our cryo-electron ...During cotranslational translocation, the signal peptide of a nascent chain binds Sec61 translocon to initiate protein transport through the endoplasmic reticulum (ER) membrane. Our cryo-electron microscopy structure of ribosome-Sec61 shows binding of an ordered heterotetrameric translocon-associated protein (TRAP) complex, in which TRAP-γ is anchored at two adjacent positions of 28 ribosomal RNA and interacts with ribosomal protein L38 and Sec61α/γ. Four transmembrane helices (TMHs) of TRAP-γ cluster with one C-terminal helix of each α, β, and δ subunits. The seven TMH bundle helps position a crescent-shaped trimeric TRAP-α/β/δ core in the ER lumen, facing the Sec61 channel. Further, our in vitro assay establishes the cyclotriazadisulfonamide derivative CK147 as a translocon inhibitor. A structure of ribosome-Sec61-CK147 reveals CK147 binding the channel and interacting with the plug helix from the lumenal side. The CK147 resistance mutations surround the inhibitor. These structures help in understanding the TRAP functions and provide a new Sec61 site for designing translocon inhibitors. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_15863.map.gz emd_15863.map.gz | 96.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-15863-v30.xml emd-15863-v30.xml emd-15863.xml emd-15863.xml | 64.8 KB 64.8 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_15863_fsc.xml emd_15863_fsc.xml | 12.4 KB | Display |  FSC data file FSC data file |
| Images |  emd_15863.png emd_15863.png | 56 KB | ||
| Others |  emd_15863_half_map_1.map.gz emd_15863_half_map_1.map.gz emd_15863_half_map_2.map.gz emd_15863_half_map_2.map.gz | 131.6 MB 131.6 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-15863 http://ftp.pdbj.org/pub/emdb/structures/EMD-15863 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-15863 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-15863 | HTTPS FTP |
-Related structure data
| Related structure data |  8b6cMC  8b5lC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_15863.map.gz / Format: CCP4 / Size: 166.4 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_15863.map.gz / Format: CCP4 / Size: 166.4 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.23 Å | ||||||||||||||||||||||||||||||||||||
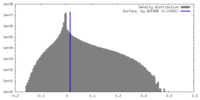
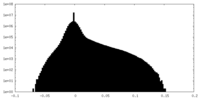
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_15863_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
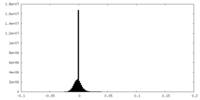
| Density Histograms |
-Half map: #1
| File | emd_15863_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
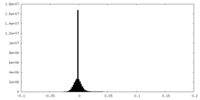
| Density Histograms |
- Sample components
Sample components
+Entire : 80S ribosome complex with Sec61 and inhibitor CK147
+Supramolecule #1: 80S ribosome complex with Sec61 and inhibitor CK147
+Macromolecule #1: 60S ribosomal protein L21
+Macromolecule #3: 60S ribosomal protein L7
+Macromolecule #4: Ribosomal_L18_c domain-containing protein
+Macromolecule #5: 60S ribosomal protein L18a
+Macromolecule #6: 60S ribosomal protein L29
+Macromolecule #7: Ribosomal protein L3
+Macromolecule #8: 60S ribosomal protein L31
+Macromolecule #10: 60S ribosomal protein L7a
+Macromolecule #11: 60S ribosomal protein L11
+Macromolecule #12: 60S ribosomal protein L30
+Macromolecule #13: Ribosomal protein L37
+Macromolecule #14: Ribosomal protein L8
+Macromolecule #15: Ribosomal protein L10
+Macromolecule #16: Protein transport protein Sec61 subunit gamma
+Macromolecule #17: 60S ribosomal protein L37a
+Macromolecule #18: 60S ribosomal protein L23
+Macromolecule #19: 60S ribosomal protein L40
+Macromolecule #20: 60S ribosomal protein L39-like
+Macromolecule #21: 60S ribosomal protein L17
+Macromolecule #22: 60S ribosomal protein L6
+Macromolecule #23: eL18
+Macromolecule #24: 60S ribosomal protein L19
+Macromolecule #25: Ribosomal protein L24
+Macromolecule #26: 60S ribosomal protein L13a
+Macromolecule #27: Protein transport protein Sec61 subunit alpha isoform 1
+Macromolecule #28: 60S ribosomal protein L4
+Macromolecule #29: 60S ribosomal protein L36a-like
+Macromolecule #30: 60S ribosomal protein L28
+Macromolecule #31: 60S ribosomal protein L35a
+Macromolecule #32: Ribosomal protein L32
+Macromolecule #33: 60S ribosomal protein L27
+Macromolecule #34: 60S ribosomal protein L27a
+Macromolecule #35: 60S ribosomal protein L34
+Macromolecule #36: 60S ribosomal protein L38
+Macromolecule #37: 60S ribosomal protein L9
+Macromolecule #38: 60S ribosomal protein L13
+Macromolecule #40: Ribosomal protein L15
+Macromolecule #41: 60S ribosomal protein L36
+Macromolecule #42: 60S ribosomal protein L35
+Macromolecule #43: 60S ribosomal protein L14
+Macromolecule #44: Ribosomal protein L26
+Macromolecule #45: Ribosomal_L23eN domain-containing protein
+Macromolecule #46: 60S ribosomal protein L22 (Fragment)
+Macromolecule #47: 60S ribosomal protein L41
+Macromolecule #2: 28S rRNA
+Macromolecule #9: 5.8S rRNA
+Macromolecule #39: 5S rRNA
+Macromolecule #48: MAGNESIUM ION
+Macromolecule #49: ZINC ION
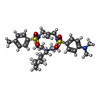
+Macromolecule #50: 4-[[9-(cyclohexylmethyl)-3-methylidene-5-(4-methylphenyl)sulfonyl...
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER / Mesh: 300 / Support film - Material: CARBON / Support film - topology: HOLEY / Support film - Film thickness: 2.0 nm / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 30 sec. / Pretreatment - Atmosphere: AIR / Pretreatment - Pressure: 0.015 kPa |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 95 % / Chamber temperature: 277.15 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | TFS GLACIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON III (4k x 4k) / Detector mode: COUNTING / Number grids imaged: 1 / Number real images: 1734 / Average exposure time: 40.0 sec. / Average electron dose: 0.8 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 50.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 1.8 µm / Nominal defocus min: 0.6 µm / Nominal magnification: 120000 |
| Sample stage | Specimen holder model: OTHER / Cooling holder cryogen: NITROGEN |
 Movie
Movie Controller
Controller










 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)