2WPM
 
 | | factor IXa superactive mutant, EGR-CMK inhibited | | Descriptor: | CALCIUM ION, COAGULATION FACTOR IXA HEAVY CHAIN, COAGULATION FACTOR IXA LIGHT CHAIN, ... | | Authors: | Zogg, T, Brandstetter, H. | | Deposit date: | 2009-08-06 | | Release date: | 2009-12-22 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Basis of the Cofactor- and Substrate-Assisted Activation of Human Coagulation Factor Ixa
Structure, 17, 2009
|
|
2WPI
 
 | | factor IXa superactive double mutant | | Descriptor: | CALCIUM ION, COAGULATION FACTOR IXA HEAVY CHAIN, COAGULATION FACTOR IXA LIGHT CHAIN, ... | | Authors: | Zogg, T, Brandstetter, H. | | Deposit date: | 2009-08-06 | | Release date: | 2009-12-22 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Structural Basis of the Cofactor- and Substrate-Assisted Activation of Human Coagulation Factor Ixa
Structure, 17, 2009
|
|
2WPK
 
 | | factor IXa superactive triple mutant, ethylene glycol-soaked | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, COAGULATION FACTOR IXA HEAVY CHAIN, ... | | Authors: | Zogg, T, Brandstetter, H. | | Deposit date: | 2009-08-06 | | Release date: | 2009-12-22 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Structural Basis of the Cofactor- and Substrate-Assisted Activation of Human Coagulation Factor Ixa
Structure, 17, 2009
|
|
2WPL
 
 | | factor IXa superactive triple mutant, EDTA-soaked | | Descriptor: | COAGULATION FACTOR IXA HEAVY CHAIN, COAGULATION FACTOR IXA LIGHT CHAIN, D-PHE-PRO-ARG-CHLOROMETHYL KETONE | | Authors: | Zogg, T, Brandstetter, H. | | Deposit date: | 2009-08-06 | | Release date: | 2009-12-22 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Structural Basis of the Cofactor- and Substrate-Assisted Activation of Human Coagulation Factor Ixa
Structure, 17, 2009
|
|
2WPJ
 
 | | factor IXa superactive triple mutant, NaCl-soaked | | Descriptor: | CALCIUM ION, COAGULATION FACTOR IXA HEAVY CHAIN, COAGULATION FACTOR IXA LIGHT CHAIN, ... | | Authors: | Zogg, T, Brandstetter, H. | | Deposit date: | 2009-08-06 | | Release date: | 2009-12-22 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural Basis of the Cofactor- and Substrate-Assisted Activation of Human Coagulation Factor Ixa
Structure, 17, 2009
|
|
2WPH
 
 | | factor IXa superactive triple mutant | | Descriptor: | CALCIUM ION, COAGULATION FACTOR IXA HEAVY CHAIN, COAGULATION FACTOR IXA LIGHT CHAIN, ... | | Authors: | Zogg, T, Brandstetter, H. | | Deposit date: | 2009-08-06 | | Release date: | 2009-12-22 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural Basis of the Cofactor- and Substrate-Assisted Activation of Human Coagulation Factor Ixa
Structure, 17, 2009
|
|
3ZFN
 
 | | Crystal structure of product-like, processed N-terminal protease Npro | | Descriptor: | CHLORIDE ION, MONOTHIOGLYCEROL, N-TERMINAL PROTEASE NPRO | | Authors: | Zogg, T, Sponring, M, Schindler, S, Koll, M, Schneider, R, Brandstetter, H, Auer, B. | | Deposit date: | 2012-12-12 | | Release date: | 2013-05-15 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal Structures of the Viral Protease Npro Imply Distinct Roles for the Catalytic Water in Catalysis
Structure, 21, 2013
|
|
3ZFQ
 
 | | Crystal structure of product-like, processed N-terminal protease Npro with mercury | | Descriptor: | MERCURY (II) ION, MONOTHIOGLYCEROL, N-TERMINAL PROTEASE NPRO | | Authors: | Zogg, T, Sponring, M, Schindler, S, Koll, M, Schneider, R, Brandstetter, H, Auer, B. | | Deposit date: | 2012-12-12 | | Release date: | 2013-05-15 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Crystal Structures of the Viral Protease Npro Imply Distinct Roles for the Catalytic Water in Catalysis
Structure, 21, 2013
|
|
3ZFT
 
 | | Crystal structure of product-like, processed N-terminal protease Npro at pH 3 | | Descriptor: | CHLORIDE ION, MONOTHIOGLYCEROL, N-TERMINAL PROTEASE NPRO | | Authors: | Zogg, T, Sponring, M, Schindler, S, Koll, M, Schneider, R, Brandstetter, H, Auer, B. | | Deposit date: | 2012-12-12 | | Release date: | 2013-05-15 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structures of the Viral Protease Npro Imply Distinct Roles for the Catalytic Water in Catalysis
Structure, 21, 2013
|
|
3ZFO
 
 | | Crystal structure of substrate-like, unprocessed N-terminal protease Npro mutant S169P | | Descriptor: | CHLORIDE ION, HYDROXIDE ION, MONOTHIOGLYCEROL, ... | | Authors: | Zogg, T, Sponring, M, Schindler, S, Koll, M, Schneider, R, Brandstetter, H, Auer, B. | | Deposit date: | 2012-12-12 | | Release date: | 2013-05-15 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Crystal Structures of the Viral Protease Npro Imply Distinct Roles for the Catalytic Water in Catalysis
Structure, 21, 2013
|
|
3ZFP
 
 | | Crystal structure of product-like, processed N-terminal protease Npro with internal His-Tag | | Descriptor: | CHLORIDE ION, MONOTHIOGLYCEROL, N-TERMINAL PROTEASE NPRO | | Authors: | Zogg, T, Sponring, M, Schindler, S, Koll, M, Schneider, R, Brandstetter, H, Auer, B. | | Deposit date: | 2012-12-12 | | Release date: | 2013-05-15 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Crystal Structures of the Viral Protease Npro Imply Distinct Roles for the Catalytic Water in Catalysis
Structure, 21, 2013
|
|
3ZFU
 
 | | Crystal structure of substrate-like, unprocessed N-terminal protease Npro mutant S169P with sulphate | | Descriptor: | MONOTHIOGLYCEROL, N-TERMINAL PROTEASE NPRO, SULFATE ION | | Authors: | Zogg, T, Sponring, M, Schindler, S, Koll, M, Schneider, R, Brandstetter, H, Auer, B. | | Deposit date: | 2012-12-12 | | Release date: | 2013-05-15 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Crystal Structures of the Viral Protease Npro Imply Distinct Roles for the Catalytic Water in Catalysis
Structure, 21, 2013
|
|
3ZFR
 
 | | Crystal structure of product-like, processed N-terminal protease Npro with iridium | | Descriptor: | HYDROXIDE ION, IRIDIUM (III) ION, MONOTHIOGLYCEROL, ... | | Authors: | Zogg, T, Sponring, M, Schindler, S, Koll, M, Schneider, R, Brandstetter, H, Auer, B. | | Deposit date: | 2012-12-12 | | Release date: | 2013-05-15 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structures of the Viral Protease Npro Imply Distinct Roles for the Catalytic Water in Catalysis
Structure, 21, 2013
|
|
6QD6
 
 | | Molecular scaffolds expand the nanobody toolkit for cryo-EM applications: crystal structure of Mb-cHopQ-Nb207 | | Descriptor: | CHLORIDE ION, Mb-cHopQ-Nb207,Outer membrane protein,Mb-cHopQ-Nb207,Outer membrane protein,Mb-cHopQ-Nb207 | | Authors: | Uchanski, T, Masiulis, S, Fischer, B, Kalichuk, V, Wohlkonig, A, Zogg, T, Remaut, H, Vranken, W, Aricescu, A.R, Pardon, E, Steyaert, J. | | Deposit date: | 2018-12-31 | | Release date: | 2019-12-18 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.84 Å) | | Cite: | Megabodies expand the nanobody toolkit for protein structure determination by single-particle cryo-EM
Nat.Methods, 18, 2021
|
|
8G8W
 
 | | Molecular mechanism of nucleotide inhibition of human uncoupling protein 1 | | Descriptor: | CARDIOLIPIN, GUANOSINE-5'-TRIPHOSPHATE, Mitochondrial brown fat uncoupling protein 1, ... | | Authors: | Gogoi, P, Jones, S.A, Ruprecht, J.J, King, M.S, Lee, Y, Zogg, T, Pardon, E, Chand, D, Steimle, S, Copeman, D, Cotrim, C.A, Steyaert, J, Crichton, P.G, Moiseenkova-Bell, V, Kunji, E.R.S. | | Deposit date: | 2023-02-20 | | Release date: | 2023-06-07 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural basis of purine nucleotide inhibition of human uncoupling protein 1.
Sci Adv, 9, 2023
|
|
6GCI
 
 | | Structure of the bongkrekic acid-inhibited mitochondrial ADP/ATP carrier | | Descriptor: | Bongkrekic acid, CARDIOLIPIN, HEXAETHYLENE GLYCOL, ... | | Authors: | Ruprecht, J.J, King, M.S, Pardon, E, Aleksandrova, A.A, Zogg, T, Crichton, P.G, Steyaert, J, Kunji, E.R.S. | | Deposit date: | 2018-04-17 | | Release date: | 2019-01-09 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | The Molecular Mechanism of Transport by the Mitochondrial ADP/ATP Carrier.
Cell, 176, 2019
|
|
9FZQ
 
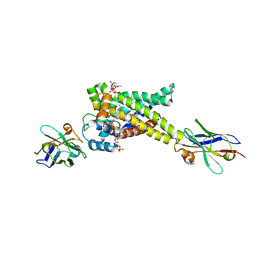 | | Proton conductance by human uncoupling protein 1 is inhibited by both purine and pyrimidine nucleotides | | Descriptor: | CA9865, CA9871, CARDIOLIPIN, ... | | Authors: | Jones, S.A, Sowton, A.P, Lacabanne, D, King, M.S, Palmer, S.M, Zogg, T, Pardon, E, Steyaert, J, Ruprecht, J.J, Kunji, E.R.S. | | Deposit date: | 2024-07-05 | | Release date: | 2025-03-05 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.03 Å) | | Cite: | Proton conductance by human uncoupling protein 1 is inhibited by purine and pyrimidine nucleotides.
Embo J., 44, 2025
|
|
5JMO
 
 | |
6QFA
 
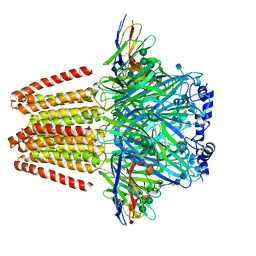 | | CryoEM structure of a beta3K279T GABA(A)R homomer in complex with histamine and megabody Mb25 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Gamma-aminobutyric acid receptor subunit beta-3,Gamma-aminobutyric acid receptor subunit beta-3, HISTAMINE, ... | | Authors: | Uchanski, T, Masiulis, S, Fischer, B, Kalichuk, V, Wohlkoening, A, Zoegg, T, Remaut, H, Vranken, W, Aricescu, A.R, Pardon, E, Steyaert, J. | | Deposit date: | 2019-01-09 | | Release date: | 2021-08-04 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (2.49 Å) | | Cite: | Megabodies expand the nanobody toolkit for protein structure determination by single-particle cryo-EM.
Nat.Methods, 18, 2021
|
|
6XVI
 
 | |
6XUX
 
 | | Crystal structure of Megabody Mb-Nb207-cYgjK_NO | | Descriptor: | CALCIUM ION, Nanobody,Glucosidase YgjK,Glucosidase YgjK,Nanobody | | Authors: | Steyaert, J, Uchanski, T, Fischer, B. | | Deposit date: | 2020-01-21 | | Release date: | 2021-01-13 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.90000641 Å) | | Cite: | Megabodies expand the nanobody toolkit for protein structure determination by single-particle cryo-EM.
Nat.Methods, 18, 2021
|
|
6XV8
 
 | |
5JMR
 
 | | X-ray structure of the furin inhibitory antibody Nb14 | | Descriptor: | CHLORIDE ION, camelid VHH fragment | | Authors: | Dahms, S.O, Than, M.E. | | Deposit date: | 2016-04-29 | | Release date: | 2016-12-07 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | The structure of a furin-antibody complex explains non-competitive inhibition by steric exclusion of substrate conformers.
Sci Rep, 6, 2016
|
|
8BE4
 
 | |
8BE5
 
 | | Crystal structure of SOS1-KRasG12V-Nanobody22-Nanobody75 | | Descriptor: | Isoform 2B of GTPase KRas, Nanobody22, Nanobody75, ... | | Authors: | Fischer, B, Wohlkonig, A, Steyaert, J. | | Deposit date: | 2022-10-21 | | Release date: | 2023-11-01 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.13 Å) | | Cite: | Allosteric nanobodies to study the interactions between SOS1 and RAS.
Nat Commun, 15, 2024
|
|
