6L2T
 
 | |
6PXG
 
 | |
6PXH
 
 | | Crystal Structure of MERS-CoV S1-NTD bound with G2 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, DIHYDROFOLIC ACID, ... | | Authors: | Wang, N, McLellan, J.S. | | Deposit date: | 2019-07-26 | | Release date: | 2019-09-25 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Definition of a Neutralization-Sensitive Epitope on the MERS-CoV S1-NTD.
Cell Rep, 28, 2019
|
|
7SPT
 
 | | Crystal structure of exofacial state human glucose transporter GLUT3 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Solute carrier family 2, facilitated glucose transporter member 3, ... | | Authors: | Wang, N, Jiang, X, Yan, N. | | Deposit date: | 2021-11-03 | | Release date: | 2022-05-18 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Molecular basis for inhibiting human glucose transporters by exofacial inhibitors.
Nat Commun, 13, 2022
|
|
7SPS
 
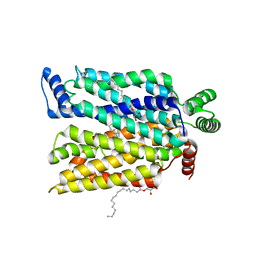 | | Crystal structure of human glucose transporter GLUT3 bound with exofacial inhibitor SA47 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Solute carrier family 2, facilitated glucose transporter member 3, ... | | Authors: | Wang, N, Jiang, X, Yan, N. | | Deposit date: | 2021-11-03 | | Release date: | 2022-05-18 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Molecular basis for inhibiting human glucose transporters by exofacial inhibitors.
Nat Commun, 13, 2022
|
|
6LYY
 
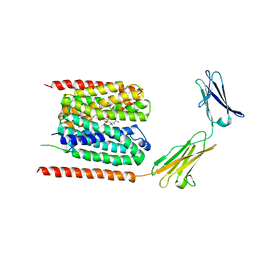 | | Cryo-EM structure of the human MCT1/Basigin-2 complex in the presence of anti-cancer drug candidate AZD3965 in the outward-open conformation. | | Descriptor: | 3-methyl-5-[[(4~{R})-4-methyl-4-oxidanyl-1,2-oxazolidin-2-yl]carbonyl]-6-[[5-methyl-3-(trifluoromethyl)-1~{H}-pyrazol-4-yl]methyl]-1-propan-2-yl-thieno[2,3-d]pyrimidine-2,4-dione, Basigin, Monocarboxylate transporter 1 | | Authors: | Wang, N, Jiang, X, Zhang, S, Zhu, A, Yuan, Y, Lei, J, Yan, C. | | Deposit date: | 2020-02-16 | | Release date: | 2020-12-23 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural basis of human monocarboxylate transporter 1 inhibition by anti-cancer drug candidates.
Cell, 184, 2021
|
|
8YQP
 
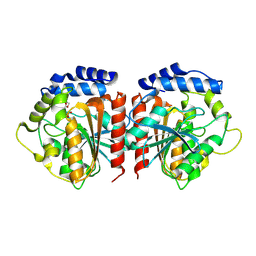 | | Crystal structure of HylD1 in complex with MEP | | Descriptor: | 2-ethoxycarbonylbenzoic acid, Lipase | | Authors: | Wang, N, Li, C.Y. | | Deposit date: | 2024-03-19 | | Release date: | 2024-07-31 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Molecular insights into the catalytic mechanism of a phthalate ester hydrolase.
J Hazard Mater, 476, 2024
|
|
8YQJ
 
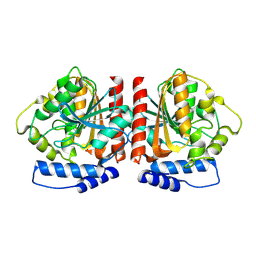 | | Crystal structure of HylD1 | | Descriptor: | Lipase | | Authors: | Wang, N, Li, C.Y. | | Deposit date: | 2024-03-19 | | Release date: | 2024-07-31 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Molecular insights into the catalytic mechanism of a phthalate ester hydrolase.
J Hazard Mater, 476, 2024
|
|
6LZ0
 
 | | Cryo-EM structure of human MCT1 in complex with Basigin-2 in the presence of lactate | | Descriptor: | (2S)-2-HYDROXYPROPANOIC ACID, Basigin, Monocarboxylate transporter 1 | | Authors: | Wang, N, Jiang, X, Zhang, S, Zhu, A, Yuan, Y, Lei, J, Yan, C. | | Deposit date: | 2020-02-16 | | Release date: | 2020-12-23 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural basis of human monocarboxylate transporter 1 inhibition by anti-cancer drug candidates.
Cell, 184, 2021
|
|
5JI8
 
 | | Crystal structure of the BRD9 bromodomain and hit 1 | | Descriptor: | 2-amino-1,3-benzothiazole-6-carboxamide, Bromodomain-containing protein 9 | | Authors: | Wang, N, Li, F, Bao, H, Li, J, Wu, J, Ruan, K. | | Deposit date: | 2016-04-22 | | Release date: | 2016-06-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | NMR Fragment Screening Hit Induces Plasticity of BRD7/9 Bromodomains
Chembiochem, 17, 2016
|
|
8Y4U
 
 | | Crystal structure of a His1 from oryza sativa | | Descriptor: | FE (III) ION, Fe(II)/2-oxoglutarate-dependent oxygenase | | Authors: | Wang, N, Ma, J.M, Shibing, H, Beibei, Y, He, Z, Dandan, L. | | Deposit date: | 2024-01-30 | | Release date: | 2024-02-28 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of HPPD inhibitor sensitive protein from Oryza sativa.
Biochem.Biophys.Res.Commun., 704, 2024
|
|
8VSP
 
 | | Cryo-EM structure of human invariant chain in complex with HLA-DQ | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, HLA class II histocompatibility antigen gamma chain, HLA class II histocompatibility antigen, ... | | Authors: | Wang, N, Caveney, N.A, Jude, K.M, Garcia, K.C. | | Deposit date: | 2024-01-24 | | Release date: | 2024-05-08 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.12 Å) | | Cite: | Structural insights into human MHC-II association with invariant chain.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8VRW
 
 | | Cryo-EM structure of human invariant chain in complex with HLA-DR15 | | Descriptor: | HLA class II histocompatibility antigen gamma chain, HLA class II histocompatibility antigen, DR alpha chain, ... | | Authors: | Wang, N, Caveney, N.A, Jude, K.M, Garcia, K.C. | | Deposit date: | 2024-01-22 | | Release date: | 2024-05-08 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.03 Å) | | Cite: | Structural insights into human MHC-II association with invariant chain.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8WWP
 
 | | PNPase mutant of Mycobacterium tuberculosis | | Descriptor: | Bifunctional guanosine pentaphosphate synthetase/polyribonucleotide nucleotidyltransferase | | Authors: | Wang, N, Sheng, Y.N, Liu, Y.T. | | Deposit date: | 2023-10-26 | | Release date: | 2024-07-03 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.12 Å) | | Cite: | Cryo-EM structures of Mycobacterium tuberculosis polynucleotide phosphorylase suggest a potential mechanism for its RNA substrate degradation.
Arch.Biochem.Biophys., 754, 2024
|
|
7EKQ
 
 | | CrClpP-S2c | | Descriptor: | ATP-dependent Clp protease ATP-binding subunit CLPT4, chloroplastic, ATP-dependent Clp protease proteolytic subunit, ... | | Authors: | Wang, N, Wang, Y.F, Cong, Y, Liu, C.M. | | Deposit date: | 2021-04-06 | | Release date: | 2021-10-20 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | The cryo-EM structure of the chloroplast ClpP complex.
Nat.Plants, 7, 2021
|
|
7EKO
 
 | | CrClpP-S1 | | Descriptor: | ATP-dependent Clp protease ATP-binding subunit CLPT4, chloroplastic, ATP-dependent Clp protease proteolytic subunit | | Authors: | Wang, N, Wang, Y.F, Cong, Y, Liu, C.M. | | Deposit date: | 2021-04-06 | | Release date: | 2021-10-20 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | The cryo-EM structure of the chloroplast ClpP complex.
Nat.Plants, 7, 2021
|
|
6J4P
 
 | |
6J4V
 
 | | Structural basis of tubulin detyrosination by vasohibins-SVBP enzyme complex and functional implications | | Descriptor: | GLYCEROL, PHOSPHATE ION, Small vasohibin-binding protein, ... | | Authors: | Wang, N, Bao, H, Huang, H. | | Deposit date: | 2019-01-10 | | Release date: | 2019-05-01 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis of tubulin detyrosination by the vasohibin-SVBP enzyme complex.
Nat.Struct.Mol.Biol., 26, 2019
|
|
8YV7
 
 | |
6J4U
 
 | |
6J4Q
 
 | | Structural basis of tubulin detyrosination by vasohibins-SVBP enzyme complex and functional implications | | Descriptor: | GLYCEROL, N-[(2S)-4-chloro-3-oxo-1-phenyl-butan-2-yl]-4-methyl-benzenesulfonamide, Small vasohibin-binding protein, ... | | Authors: | Wang, N, Bao, H, Huang, H. | | Deposit date: | 2019-01-10 | | Release date: | 2019-05-01 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis of tubulin detyrosination by the vasohibin-SVBP enzyme complex.
Nat.Struct.Mol.Biol., 26, 2019
|
|
6J4O
 
 | |
6J4S
 
 | | Structural basis of tubulin detyrosination by vasohibins-SVBP enzyme complex and functional implications | | Descriptor: | GLYCEROL, PHOSPHATE ION, Small vasohibin-binding protein, ... | | Authors: | Wang, N, Bao, H, Huang, H. | | Deposit date: | 2019-01-10 | | Release date: | 2019-05-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis of tubulin detyrosination by the vasohibin-SVBP enzyme complex.
Nat.Struct.Mol.Biol., 26, 2019
|
|
4I9O
 
 | | Crystal Structure of GACKIX L664C Tethered to 1-10 | | Descriptor: | 1,2-ETHANEDIOL, 1-{4-[4-chloro-3-(trifluoromethyl)phenyl]-4-hydroxypiperidin-1-yl}-3-sulfanylpropan-1-one, CREB-binding protein | | Authors: | Wang, N, Meagher, J.L, Stuckey, J.A, Mapp, A.K. | | Deposit date: | 2012-12-05 | | Release date: | 2013-03-06 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Ordering a dynamic protein via a small-molecule stabilizer.
J.Am.Chem.Soc., 135, 2013
|
|
6A5W
 
 | | FXR-LBD with HNC143 and SRC1 | | Descriptor: | 2-[2-[[3-[2,6-bis(chloranyl)phenyl]-5-cyclopropyl-1,2-oxazol-4-yl]methoxy]-6-azaspiro[3.4]octan-6-yl]-1,3-benzothiazole-6-carboxylic acid, Bile acid receptor, Nuclear receptor coactivator 1 | | Authors: | Wang, N, Liu, J. | | Deposit date: | 2018-06-25 | | Release date: | 2018-10-10 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.88 Å) | | Cite: | Ligand binding and heterodimerization with retinoid X receptor alpha (RXR alpha ) induce farnesoid X receptor (FXR) conformational changes affecting coactivator binding
J. Biol. Chem., 293, 2018
|
|
