2KDI
 
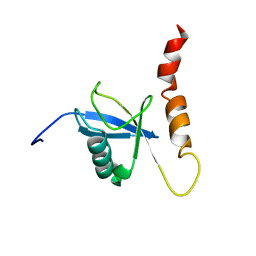 | | Solution structure of a Ubiquitin/UIM fusion protein | | Descriptor: | Ubiquitin, Vacuolar protein sorting-associated protein 27 fusion protein | | Authors: | Sgourakis, N.G, Patel, M.M, Garcia, A.E, Makhatadze, G.I, McCallum, S.A. | | Deposit date: | 2009-01-09 | | Release date: | 2010-02-09 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Conformational Dynamics and Structural Plasticity Play Critical Roles in the Ubiquitin Recognition of a UIM Domain.
J.Mol.Biol., 396, 2010
|
|
2MPZ
 
 | |
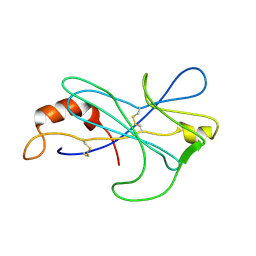
2MIZ
 
 | |
3J1W
 
 | |
3J1X
 
 | | A refined model of the prototypical Salmonella typhimurium T3SS basal body reveals the molecular basis for its assembly | | Descriptor: | Protein PrgH | | Authors: | Sgourakis, N.G, Bergeron, J.R.C, Worrall, L.J, Strynadka, N.C.J, Baker, D. | | Deposit date: | 2012-07-10 | | Release date: | 2013-05-22 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (11.7 Å) | | Cite: | A Refined Model of the Prototypical Salmonella SPI-1 T3SS Basal Body Reveals the Molecular Basis for Its Assembly.
Plos Pathog., 9, 2013
|
|
3J1V
 
 | |
5TXS
 
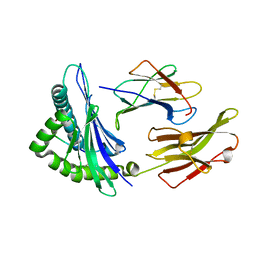 | | Crystal structure of an anaplastic lymphoma kinase-derived neuroblastoma tumor antigen bound to the Human Major Histocompatibility Complex Class I molecule HLA-B*1501 | | Descriptor: | Beta-2-microglobulin, HLA class I histocompatibility antigen, B-15 alpha chain, ... | | Authors: | Toor, J, Rao, A.A, Salama, S, Tripathi, S, Haussler, D, Sgourakis, N.G. | | Deposit date: | 2016-11-17 | | Release date: | 2017-11-29 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.697 Å) | | Cite: | A Recurrent Mutation in Anaplastic Lymphoma Kinase with Distinct Neoepitope Conformations.
Front Immunol, 9, 2018
|
|
6OSW
 
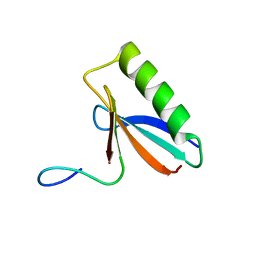 | | An order-to-disorder structural switch activates the FoxM1 transcription factor | | Descriptor: | Forkhead box M1 | | Authors: | Marceau, A.H, Rubin, S.M, Nerli, S, McShane, A.C, Sgourakis, N.G. | | Deposit date: | 2019-05-02 | | Release date: | 2019-05-29 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | An order-to-disorder structural switch activates the FoxM1 transcription factor.
Elife, 8, 2019
|
|
9C96
 
 | | Cryo-EM structure of TAP binding protein related (TAPBPR) in complex with HLA-A*02:01 bound to a suboptimal peptide. | | Descriptor: | Beta-2-microglobulin, LYS-ILE-LEU-GLY-PHE-VAL, MHC class I antigen, ... | | Authors: | Pumroy, R.P, Mallik, L, Sun, Y, Moiseenkova-Bell, Y.V, Sgourakis, N.G. | | Deposit date: | 2024-06-13 | | Release date: | 2025-01-22 | | Last modified: | 2025-01-29 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | CryoEM structure of an MHC-I/TAPBPR peptide-bound intermediate reveals the mechanism of antigen proofreading.
Proc.Natl.Acad.Sci.USA, 122, 2025
|
|
8SBK
 
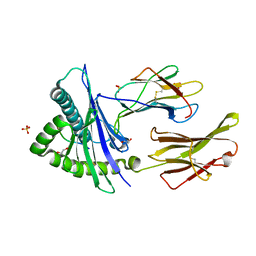 | | Structure of HLA-A*24:02 in complex with peptide, LYLPVRVLI (ATG2A). | | Descriptor: | 1,2-ETHANEDIOL, Beta-2-microglobulin, LEU-TYR-LEU-PRO-VAL-ARG-VAL-LEU-ILE, ... | | Authors: | Mallik, L, Young, M.C, Sgourakis, N.G. | | Deposit date: | 2023-04-03 | | Release date: | 2023-12-06 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural principles of peptide-centric chimeric antigen receptor recognition guide therapeutic expansion.
Sci Immunol, 8, 2023
|
|
8SBL
 
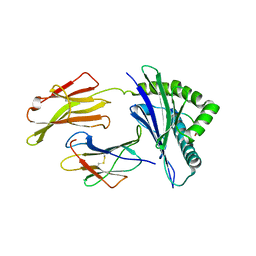 | | Structure of HLA-A*24:02 in complex with peptide, LYLPVRVLI | | Descriptor: | Beta-2-microglobulin, LEU-TYR-LEU-PRO-VAL-ARG-VAL-LEU-ILE, MHC class I antigen | | Authors: | Mallik, L, Young, M.C, Sgourakis, N.G. | | Deposit date: | 2023-04-03 | | Release date: | 2023-12-06 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural principles of peptide-centric chimeric antigen receptor recognition guide therapeutic expansion.
Sci Immunol, 8, 2023
|
|
6AT9
 
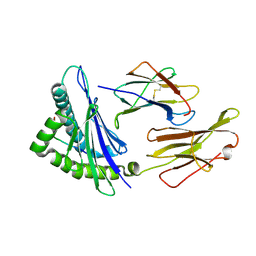 | | Crystal structure of an anaplastic lymphoma kinase-derived neuroblastoma tumor antigen bound to the Human Major Histocompatibility Complex Class I molecule HLA-A*0101 | | Descriptor: | ALK, Beta-2-microglobulin, HLA class I histocompatibility antigen, ... | | Authors: | Toor, J, Rao, A.A, Salama, S, Tripathi, S, Haussler, D, Sgourakis, N.G. | | Deposit date: | 2017-08-28 | | Release date: | 2018-03-21 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.9503 Å) | | Cite: | A Recurrent Mutation in Anaplastic Lymphoma Kinase with Distinct Neoepitope Conformations.
Front Immunol, 9, 2018
|
|
8EK5
 
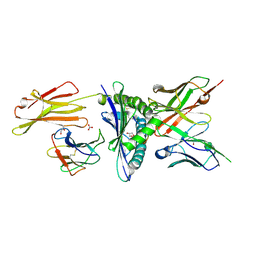 | | Engineered scFv 10LH bound to PHOX2B/HLA-A24:02 | | Descriptor: | 10LH single chain fragment variable (scFv), Beta-2-microglobulin, GLYCEROL, ... | | Authors: | Garfinkle, S.E, Florio, T.J, Sgourakis, N.G. | | Deposit date: | 2022-09-20 | | Release date: | 2023-12-06 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Structural principles of peptide-centric chimeric antigen receptor recognition guide therapeutic expansion.
Sci Immunol, 8, 2023
|
|
9DL1
 
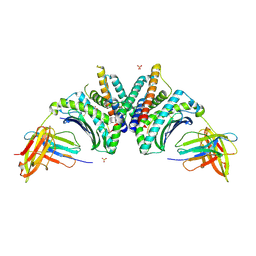 | | Crystal Structure of HLA-A*02:01/NY-ESO-1 (SLLMWITQV) and a target specific TRACeR-I | | Descriptor: | Beta-2-microglobulin, Cancer/testis antigen 1, MHC class I antigen, ... | | Authors: | Mallik, L, Du, H, Huang, P, Sgourakis, N.G. | | Deposit date: | 2024-09-10 | | Release date: | 2024-11-20 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Targeting peptide antigens using a multiallelic MHC I-binding system.
Nat.Biotechnol., 2024
|
|
8ESH
 
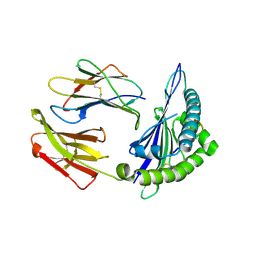 | | Structure of chimeric HLA-A*02:01 bound to CMV peptide | | Descriptor: | Beta-2-microglobulin, CMV peptide, HLA-A*02:01 | | Authors: | Florio, T.J, Ani, O, Young, M.C, Mallik, L, Sgourakis, N.G. | | Deposit date: | 2022-10-14 | | Release date: | 2023-01-25 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.72 Å) | | Cite: | Decoupling peptide binding from T cell receptor recognition with engineered chimeric MHC-I molecules.
Front Immunol, 14, 2023
|
|
8ERX
 
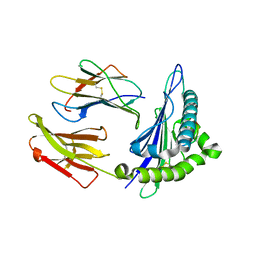 | | Structure of chimeric HLA-A*11:01-A*02:01 bound to HIV-1 RT peptide | | Descriptor: | Beta-2-microglobulin, HIV-1 RT, HLA-A*02:01 | | Authors: | Florio, T.J, Ani, O, Young, M.C, Mallik, L, Sgourakis, N.G. | | Deposit date: | 2022-10-13 | | Release date: | 2023-01-25 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Decoupling peptide binding from T cell receptor recognition with engineered chimeric MHC-I molecules.
Front Immunol, 14, 2023
|
|
5WOX
 
 | |
5WOZ
 
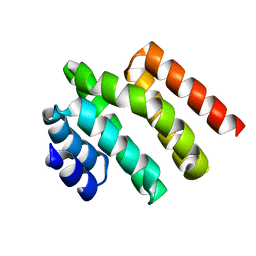 | |
5WOT
 
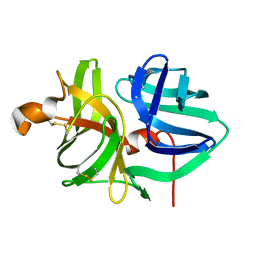 | |
5WOY
 
 | |
5VZ5
 
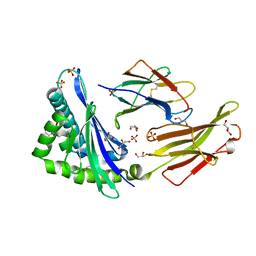 | | Crystal structure of an anaplastic lymphoma kinase-derived neuroblastoma tumor antigen bound to the Human Major Histocompatibility Complex Class I molecule HLA-B*1501 | | Descriptor: | Beta-2-microglobulin, GLYCEROL, HLA class I histocompatibility antigen, ... | | Authors: | Toor, J, Rao, A.A, Salama, S, Tripathi, S, Haussler, D, Sgourakis, N.G. | | Deposit date: | 2017-05-26 | | Release date: | 2018-03-21 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.5901 Å) | | Cite: | A Recurrent Mutation in Anaplastic Lymphoma Kinase with Distinct Neoepitope Conformations.
Front Immunol, 9, 2018
|
|
6DMP
 
 | | De Novo Design of a Protein Heterodimer with Specificity Mediated by Hydrogen Bond Networks | | Descriptor: | Designed orthogonal protein DHD13_XAAA_A, Designed orthogonal protein DHD13_XAAA_B | | Authors: | Chen, Z, Flores-Solis, D, Sgourakis, N.G, Baker, D. | | Deposit date: | 2018-06-05 | | Release date: | 2018-12-19 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Programmable design of orthogonal protein heterodimers.
Nature, 565, 2019
|
|
7MJ9
 
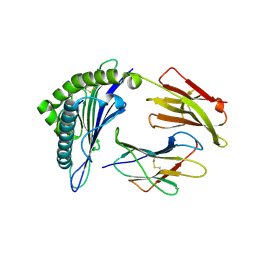 | | HLA-A*02:01 bound to Neuroblastoma Derived mutant IGFBPL1 peptide | | Descriptor: | Beta-2-microglobulin, Insulin-like growth factor-binding protein-like 1 altered peptide, MHC class I antigen | | Authors: | Toor, J.S, Tripathi, S.M, Truong, H.V, Yarmarkovich, M, Maris, J.M, Sgourakis, N.G. | | Deposit date: | 2021-04-19 | | Release date: | 2021-08-18 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Cross-HLA targeting of intracellular oncoproteins with peptide-centric CARs.
Nature, 599, 2021
|
|
7MJ6
 
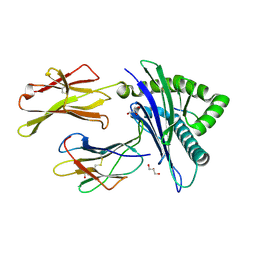 | | HLA-A*02:01 bound to Neuroblastoma Derived IGFBPL1 peptide | | Descriptor: | Beta-2-microglobulin, GLYCEROL, Insulin-like growth factor-binding protein-like 1 peptide, ... | | Authors: | Toor, J.S, Tripathi, S.M, Truong, H.V, Yarmarkovich, M, Maris, J.M, Sgourakis, N.G. | | Deposit date: | 2021-04-19 | | Release date: | 2021-08-18 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Cross-HLA targeting of intracellular oncoproteins with peptide-centric CARs.
Nature, 599, 2021
|
|
7MJ7
 
 | | HLA-A*02:01 bound to Neuroblastoma Derived IGFBPL1 peptide | | Descriptor: | Beta-2-microglobulin, GLYCEROL, Insulin-like growth factor-binding protein-like 1 peptide, ... | | Authors: | Toor, J.S, Tripathi, S.M, Truong, H.V, Yarmarkovich, M, Maris, J.M, Sgourakis, N.G. | | Deposit date: | 2021-04-19 | | Release date: | 2021-08-18 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Cross-HLA targeting of intracellular oncoproteins with peptide-centric CARs.
Nature, 599, 2021
|
|
