3J9M
 
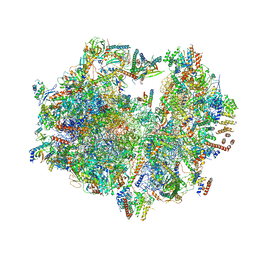 | | Structure of the human mitochondrial ribosome (class 1) | | Descriptor: | 12S rRNA, 16S rRNA, E-site tRNA, ... | | Authors: | Amunts, A, Brown, A, Toots, J, Scheres, S.H, Ramakrishnan, V. | | Deposit date: | 2015-02-08 | | Release date: | 2015-04-15 | | Last modified: | 2018-07-18 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Ribosome. The structure of the human mitochondrial ribosome.
Science, 348, 2015
|
|
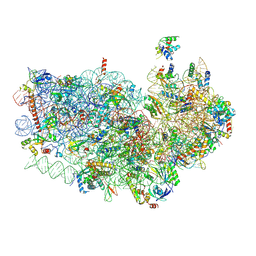
6OKK
 
 | |
5UMD
 
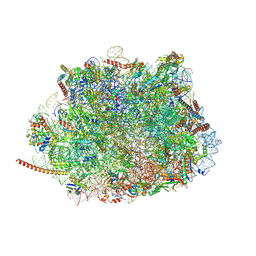 | | Structure of the Plasmodium falciparum 80S ribosome bound to the antimalarial drug mefloquine | | Descriptor: | 28S ribosomal RNA, 5.8S ribosomal RNA, 5S ribosomal RNA, ... | | Authors: | Wong, W, Bai, X.-C, Brown, A, Scheres, S, Baum, J. | | Deposit date: | 2017-01-27 | | Release date: | 2017-03-01 | | Last modified: | 2019-12-11 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Mefloquine targets the Plasmodium falciparum 80S ribosome to inhibit protein synthesis.
Nat Microbiol, 2, 2017
|
|
5GAO
 
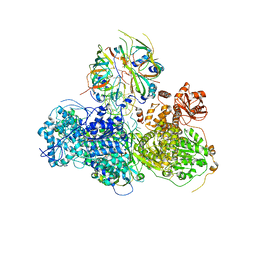 | | Head region of the yeast spliceosomal U4/U6.U5 tri-snRNP | | Descriptor: | Pre-mRNA-splicing factor 8, Pre-mRNA-splicing helicase BRR2, Saccharomyces cerevisiae strain UOA_M2 chromosome 5 sequence, ... | | Authors: | Nguyen, T.H.D, Galej, W.P, Bai, X.C, Oubridge, C, Scheres, S.H.W, Newman, A.J, Nagai, K. | | Deposit date: | 2015-12-15 | | Release date: | 2016-01-27 | | Last modified: | 2019-10-02 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Cryo-EM structure of the yeast U4/U6.U5 tri-snRNP at 3.7 angstrom resolution.
Nature, 530, 2016
|
|
5GAN
 
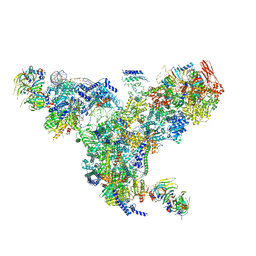 | | The overall structure of the yeast spliceosomal U4/U6.U5 tri-snRNP at 3.7 Angstrom | | Descriptor: | 13 kDa ribonucleoprotein-associated protein, GUANOSINE-5'-TRIPHOSPHATE, Pre-mRNA-processing factor 31, ... | | Authors: | Nguyen, T.H.D, Galej, W.P, Bai, X.C, Oubridge, C, Scheres, S.H.W, Newman, A.J, Nagai, K. | | Deposit date: | 2015-12-15 | | Release date: | 2016-01-27 | | Last modified: | 2019-12-11 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Cryo-EM structure of the yeast U4/U6.U5 tri-snRNP at 3.7 angstrom resolution.
Nature, 530, 2016
|
|
5M1S
 
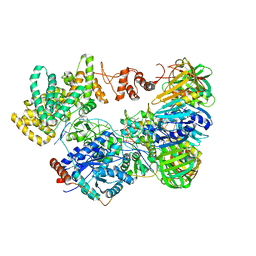 | | Cryo-EM structure of the E. coli replicative DNA polymerase-clamp-exonuclase-theta complex bound to DNA in the editing mode | | Descriptor: | DNA Primer Strand, DNA Template Strand, DNA polymerase III subunit alpha, ... | | Authors: | Fernandez-Leiro, R, Conrad, J, Scheres, S.H.W, Lamers, M.H. | | Deposit date: | 2016-10-10 | | Release date: | 2017-01-18 | | Last modified: | 2018-10-24 | | Method: | ELECTRON MICROSCOPY (6.7 Å) | | Cite: | Self-correcting mismatches during high-fidelity DNA replication.
Nat. Struct. Mol. Biol., 24, 2017
|
|
5MZ6
 
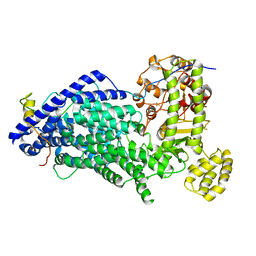 | | Cryo-EM structure of a Separase-Securin complex from Caenorhabditis elegans at 3.8 A resolution | | Descriptor: | Interactor of FizzY protein, SEParase | | Authors: | Boland, A, Martin, T.G, Zhang, Z, Yang, J, Bai, X.C, Chang, L, Scheres, S.H.W, Barford, D. | | Deposit date: | 2017-01-31 | | Release date: | 2017-03-08 | | Last modified: | 2019-12-11 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Cryo-EM structure of a metazoan separase-securin complex at near-atomic resolution.
Nat. Struct. Mol. Biol., 24, 2017
|
|
6QJP
 
 | | Cryo-EM structure of heparin-induced 2N4R tau jagged filaments | | Descriptor: | Microtubule-associated protein tau | | Authors: | Zhang, W, Falcon, B, Murzin, A.G, Fan, J, Crowther, R.A, Goedert, M, Scheres, S.H.W. | | Deposit date: | 2019-01-24 | | Release date: | 2019-02-20 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Heparin-induced tau filaments are polymorphic and differ from those in Alzheimer's and Pick's diseases.
Elife, 8, 2019
|
|
6QJH
 
 | | Cryo-EM structure of heparin-induced 2N4R tau snake filaments | | Descriptor: | Microtubule-associated protein tau | | Authors: | Zhang, W, Falcon, B, Murzin, A.G, Fan, J, Crowther, R.A, Goedert, M, Scheres, S.H.W. | | Deposit date: | 2019-01-24 | | Release date: | 2019-02-20 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Heparin-induced tau filaments are polymorphic and differ from those in Alzheimer's and Pick's diseases.
Elife, 8, 2019
|
|
6QJM
 
 | | Cryo-EM structure of heparin-induced 2N4R tau twister filaments | | Descriptor: | Microtubule-associated protein tau | | Authors: | Zhang, W, Falcon, B, Murzin, A.G, Fan, J, Crowther, R.A, Goedert, M, Scheres, S.H.W. | | Deposit date: | 2019-01-24 | | Release date: | 2019-02-27 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Heparin-induced tau filaments are polymorphic and differ from those in Alzheimer's and Pick's diseases.
Elife, 8, 2019
|
|
6QJQ
 
 | | Cryo-EM structure of heparin-induced 2N3R tau filaments | | Descriptor: | Microtubule-associated protein tau | | Authors: | Zhang, W, Falcon, B, Murzin, A.G, Fan, J, Crowther, R.A, Goedert, M, Scheres, S.H.W. | | Deposit date: | 2019-01-24 | | Release date: | 2019-02-20 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Heparin-induced tau filaments are polymorphic and differ from those in Alzheimer's and Pick's diseases.
Elife, 8, 2019
|
|
6RZA
 
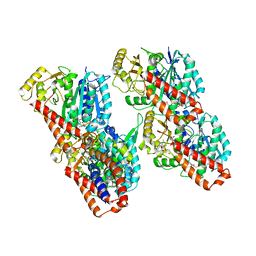 | | Cryo-EM structure of the human inner arm dynein DNAH7 microtubule binding domain bound to microtubules | | Descriptor: | Cytoplasmic dynein 1 heavy chain 1,Dynein heavy chain 7, axonemal,Cytoplasmic dynein 1 heavy chain 1, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Lacey, S.E, He, S, Scheres, S.H.W, Carter, A.P. | | Deposit date: | 2019-06-13 | | Release date: | 2019-07-10 | | Last modified: | 2019-07-17 | | Method: | ELECTRON MICROSCOPY (5.4 Å) | | Cite: | Cryo-EM of dynein microtubule-binding domains shows how an axonemal dynein distorts the microtubule.
Elife, 8, 2019
|
|
6RZB
 
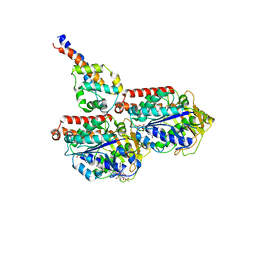 | | Cryo-EM structure of mouse cytoplasmic dynein-1 microtubule binding domain bound to microtubules | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Lacey, S.E, He, S, Scheres, S.H.W, Carter, A.P. | | Deposit date: | 2019-06-13 | | Release date: | 2019-07-10 | | Last modified: | 2019-07-17 | | Method: | ELECTRON MICROSCOPY (5 Å) | | Cite: | Cryo-EM of dynein microtubule-binding domains shows how an axonemal dynein distorts the microtubule.
Elife, 8, 2019
|
|
5GAP
 
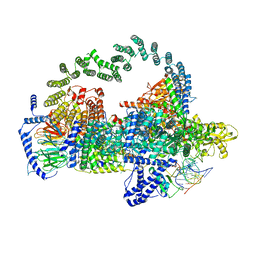 | | Body region of the U4/U6.U5 tri-snRNP | | Descriptor: | 13 kDa ribonucleoprotein-associated protein, Pre-mRNA-processing factor 31, Pre-mRNA-splicing factor 6, ... | | Authors: | Nguyen, T.H.D, Galej, W.P, Oubridge, C, Bai, X.C, Newman, A, Scheres, S, Nagai, K. | | Deposit date: | 2015-12-15 | | Release date: | 2016-01-27 | | Last modified: | 2019-10-02 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Cryo-EM structure of the yeast U4/U6.U5 tri-snRNP at 3.7 angstrom resolution.
Nature, 530, 2016
|
|
5FKU
 
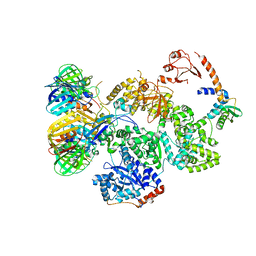 | | cryo-EM structure of the E. coli replicative DNA polymerase complex in DNA free state (DNA polymerase III alpha, beta, epsilon, tau complex) | | Descriptor: | DNA POLYMERASE III SUBUNIT ALPHA, DNA POLYMERASE III SUBUNIT BETA, DNA POLYMERASE III SUBUNIT EPSILON, ... | | Authors: | Fernandez-Leiro, R, Conrad, J, Scheres, S.H.W, Lamers, M.H. | | Deposit date: | 2015-10-20 | | Release date: | 2015-11-25 | | Last modified: | 2019-05-08 | | Method: | ELECTRON MICROSCOPY (8.34 Å) | | Cite: | cryo-EM structures of theE. colireplicative DNA polymerase reveal its dynamic interactions with the DNA sliding clamp, exonuclease andtau.
Elife, 4, 2015
|
|
5FN3
 
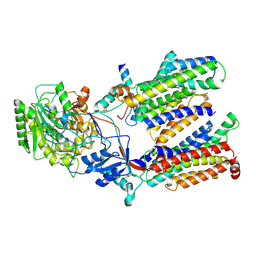 | | Cryo-EM structure of gamma secretase in class 1 of the apo- state ensemble | | Descriptor: | Gamma-secretase subunit APH-1A, Gamma-secretase subunit PEN-2, Nicastrin, ... | | Authors: | Bai, X.C, Rajendra, E, Yang, G.H, Shi, Y.G, Scheres, S.H.W. | | Deposit date: | 2015-11-10 | | Release date: | 2015-12-16 | | Last modified: | 2019-09-11 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Sampling the conformational space of the catalytic subunit of human gamma-secretase.
Elife, 4, 2015
|
|
5FN2
 
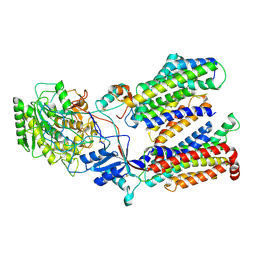 | | Cryo-EM structure of gamma secretase in complex with a drug DAPT | | Descriptor: | Gamma-secretase subunit APH-1A, Gamma-secretase subunit PEN-2, Nicastrin, ... | | Authors: | Bai, X.C, Rajendra, E, Yang, G.H, Shi, Y.G, Scheres, S.H.W. | | Deposit date: | 2015-11-10 | | Release date: | 2015-12-16 | | Last modified: | 2019-09-04 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Sampling the conformational space of the catalytic subunit of human gamma-secretase.
Elife, 4, 2015
|
|
5FN4
 
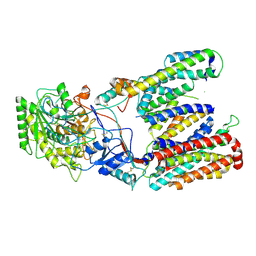 | | Cryo-EM structure of gamma secretase in class 2 of the apo- state ensemble | | Descriptor: | Gamma-secretase subunit APH-1A, Gamma-secretase subunit PEN-2, Nicastrin, ... | | Authors: | Bai, X.C, Rajendra, E, Yang, G.H, Shi, Y.G, Scheres, S.H.W. | | Deposit date: | 2015-11-10 | | Release date: | 2015-12-16 | | Last modified: | 2019-09-11 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Sampling the conformational space of the catalytic subunit of human gamma-secretase.
Elife, 4, 2015
|
|
5FKW
 
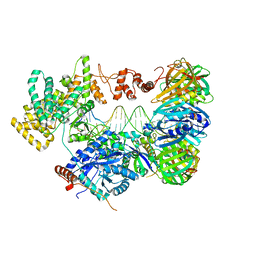 | | cryo-EM structure of the E. coli replicative DNA polymerase complex bound to DNA (DNA polymerase III alpha, beta, epsilon) | | Descriptor: | DNA POLYMERASE III ALPHA, DNA POLYMERASE III BETA, DNA POLYMERASE III EPSILON, ... | | Authors: | Fernandez-Leiro, R, Conrad, J, Scheres, S.H.W, Lamers, M.H. | | Deposit date: | 2015-10-20 | | Release date: | 2015-11-25 | | Last modified: | 2019-02-27 | | Method: | ELECTRON MICROSCOPY (7.3 Å) | | Cite: | cryo-EM structures of theE. colireplicative DNA polymerase reveal its dynamic interactions with the DNA sliding clamp, exonuclease andtau.
Elife, 4, 2015
|
|
5FN5
 
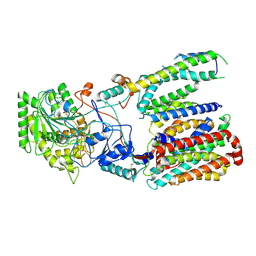 | | Cryo-EM structure of gamma secretase in class 3 of the apo- state ensemble | | Descriptor: | Gamma-secretase subunit APH-1A, Gamma-secretase subunit PEN-2, Nicastrin, ... | | Authors: | Bai, X.C, Rajendra, E, Yang, G.H, Shi, Y.G, Scheres, S.H.W. | | Deposit date: | 2015-11-10 | | Release date: | 2015-12-16 | | Last modified: | 2019-09-11 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Sampling the conformational space of the catalytic subunit of human gamma-secretase.
Elife, 4, 2015
|
|
5FKV
 
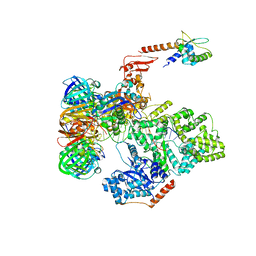 | | cryo-EM structure of the E. coli replicative DNA polymerase complex bound to DNA (DNA polymerase III alpha, beta, epsilon, tau complex) | | Descriptor: | DNA POLYMERASE III BETA, DNA POLYMERASE III EPSILON, DNA POLYMERASE III SUBUNIT ALPHA, ... | | Authors: | Fernandez-Leiro, R, Conrad, J, Scheres, S.H.W, Lamers, M.H. | | Deposit date: | 2015-10-20 | | Release date: | 2015-11-25 | | Last modified: | 2019-02-27 | | Method: | ELECTRON MICROSCOPY (8 Å) | | Cite: | cryo-EM structures of theE. colireplicative DNA polymerase reveal its dynamic interactions with the DNA sliding clamp, exonuclease andtau.
Elife, 4, 2015
|
|
8G8F
 
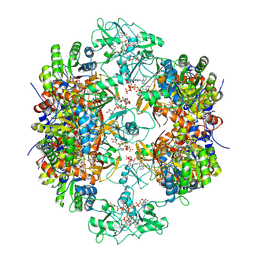 | | Human IMPDH2 mutant - L245P, treated with ATP, IMP, and NAD+; extended filament segment reconstruction | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, INOSINIC ACID, Inosine-5'-monophosphate dehydrogenase 2, ... | | Authors: | O'Neill, A.G, Kollman, J.M. | | Deposit date: | 2023-02-17 | | Release date: | 2023-04-19 | | Last modified: | 2023-08-09 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Neurodevelopmental disorder mutations in the purine biosynthetic enzyme IMPDH2 disrupt its allosteric regulation.
J.Biol.Chem., 299, 2023
|
|
8G9B
 
 | | Human IMPDH2 mutant - L245P, treated with GTP, ATP, IMP, and NAD+; compressed filament segment reconstruction | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, INOSINIC ACID, ... | | Authors: | O'Neill, A.G, Kollman, J.M. | | Deposit date: | 2023-02-21 | | Release date: | 2023-04-19 | | Last modified: | 2023-08-09 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Neurodevelopmental disorder mutations in the purine biosynthetic enzyme IMPDH2 disrupt its allosteric regulation.
J.Biol.Chem., 299, 2023
|
|
5LJW
 
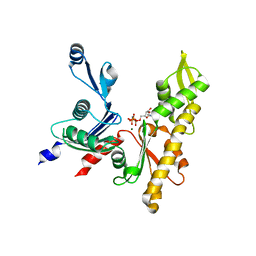 | | MamK non-polymerising A278D mutant bound to AMPPNP | | Descriptor: | Actin-like ATPase, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Lowe, J. | | Deposit date: | 2016-07-20 | | Release date: | 2016-11-16 | | Last modified: | 2017-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | X-ray and cryo-EM structures of monomeric and filamentous actin-like protein MamK reveal changes associated with polymerization.
Proc. Natl. Acad. Sci. U.S.A., 113, 2016
|
|
5LJV
 
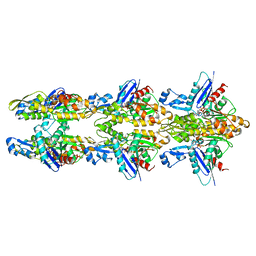 | | MamK double helical filament | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin-like ATPase, MAGNESIUM ION | | Authors: | Lowe, J. | | Deposit date: | 2016-07-21 | | Release date: | 2016-11-16 | | Last modified: | 2019-10-23 | | Method: | ELECTRON MICROSCOPY (3.65 Å) | | Cite: | X-ray and cryo-EM structures of monomeric and filamentous actin-like protein MamK reveal changes associated with polymerization.
Proc. Natl. Acad. Sci. U.S.A., 113, 2016
|
|
