3KQK
 
 | |
3KQH
 
 | |
3KQL
 
 | |
3KQN
 
 | |
3KQU
 
 | |
1ZH1
 
 | |
1A1R
 
 | | HCV NS3 PROTEASE DOMAIN:NS4A PEPTIDE COMPLEX | | Descriptor: | NS3 PROTEIN, NS4A PROTEIN, ZINC ION | | Authors: | Kim, J.L, Morgenstern, K.A, Lin, C, Fox, T, Dwyer, M.D, Landro, J.A, Chambers, S.P, Markland, W, Lepre, C.A, O'Malley, E.T, Harbeson, S.L, Rice, C.M, Murcko, M.A, Caron, P.R, Thomson, J.A. | | Deposit date: | 1997-12-15 | | Release date: | 1998-06-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of the hepatitis C virus NS3 protease domain complexed with a synthetic NS4A cofactor peptide.
Cell(Cambridge,Mass.), 87, 1996
|
|
4J1V
 
 | | Functional and structural studies of MOBKL1B, a Salvador/Warts/Hippo tumor suppressor pathway, in HCV replication | | Descriptor: | MOB kinase activator 1A, NS5A domain II peptide, ZINC ION | | Authors: | Chung, H.-Y, Gu, M, Rice, C.M. | | Deposit date: | 2013-02-02 | | Release date: | 2014-08-06 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Seed Sequence-Matched Controls Reveal Limitations of Small Interfering RNA Knockdown in Functional and Structural Studies of Hepatitis C Virus NS5A-MOBKL1B Interaction.
J.Virol., 88, 2014
|
|
5E4F
 
 | |
2HD0
 
 | |
6VJT
 
 | |
8E7M
 
 | |
2A4G
 
 | | Hepatitis C Protease NS3-4A serine protease with Ketoamide Inhibitor SCH225724 Bound | | Descriptor: | ({1-[1-CARBAMOYL-PHENYL-METHYL)-CARBAMOYL]-METHYL}-AMINOOXALYL)-BUTYLCARBAMOYL)-3-METHYL-BUTYLCARBAMOYL)-CYCLOHEXYL-METHYL)-CARBAMIC ACID ISOBUTYL ESTER, NS3 protease/helicase, NS4a peptide, ... | | Authors: | Arasappan, A, Njoroge, F.G, Chan, T.Y, Bennett, F, Bogen, S.L, Chen, K, Gu, H, Hong, L, Jao, E, Liu, Y.T, Lovey, R.G, Parekh, T, Pike, R.E, Pinto, P, Santhanam, B, Venkatraman, S, Vaccaro, H, Wang, H, Yang, X, Zhu, Z, Mckittrick, B, Saksena, A.K, Girijavallabhan, V, Pichardo, J, Butkiewicz, N, Ingram, R, Malcolm, B, Prongay, A.J, Yao, N, Marten, B, Madison, V, Kemp, S, Levy, O, Lim-Wilby, M, Tamura, S, Ganguly, A.K. | | Deposit date: | 2005-06-28 | | Release date: | 2006-07-04 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Hepatitis C virus NS3-4a serine protease inhibitors. SAR of P2' moiety with improved potency.
Bioorg.Med.Chem.Lett., 15, 2005
|
|
2A4R
 
 | | HCV NS3 Protease Domain with a Ketoamide Inhibitor Covalently bound. | | Descriptor: | NS3 protease/helicase, Ns4a peptide, ZINC ION, ... | | Authors: | Bogen, S, Saksena, A.K, Arasappan, A, Gu, H, Njoroge, F.G, Girijavallabhan, V, Pichardo, J, Butkiewicz, N, Prongay, A, Madison, V. | | Deposit date: | 2005-06-29 | | Release date: | 2006-07-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Hepatitis C Virus NS3-4A serine protease inhibitors: Use of a P2-P1 cyclopropyl alanine combination for improved potency.
Bioorg.Med.Chem.Lett., 15, 2005
|
|
2A4Q
 
 | | HCV NS3 protease with NS4a peptide and a covalently bound macrocyclic ketoamide compound. | | Descriptor: | (2R)-({N-[(3S)-3-({[(3S,6S)-6-CYCLOHEXYL-5,8-DIOXO-4,7-DIAZABICYCLO[14.3.1]ICOSA-1(20),16,18-TRIEN-3-YL]CARBONYL}AMINO)-2-OXOHEXANOYL]GLYCYL}AMINO)(PHENYL)ACETIC ACID, BETA-MERCAPTOETHANOL, NS3 protease/helicase', ... | | Authors: | Chen, K.X, Njoroge, F.G, Prongay, A, Pichardo, J, Madison, V, Girijavallabhan, V. | | Deposit date: | 2005-06-29 | | Release date: | 2006-07-04 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Synthesis and Biological Activity of Macrocyclic Inhibitors of Hepatitis C Virus (HCV) NS3 Protease
Bioorg.Med.Chem.Lett., 15, 2005
|
|
3P5O
 
 | | Crystal Structure of the First Bromodomain of Human Brd4 in complex with IBET inhibitor | | Descriptor: | 1,2-ETHANEDIOL, 2-[(4S)-6-(4-chlorophenyl)-8-methoxy-1-methyl-4H-[1,2,4]triazolo[4,3-a][1,4]benzodiazepin-4-yl]-N-ethylacetamide, Bromodomain-containing protein 4 | | Authors: | Chung, C. | | Deposit date: | 2010-10-09 | | Release date: | 2010-11-17 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Suppression of inflammation by a synthetic histone mimic
Nature, 468, 2010
|
|
1BT7
 
 | | THE SOLUTION NMR STRUCTURE OF THE N-TERMINAL PROTEASE DOMAIN OF THE HEPATITIS C VIRUS (HCV) NS3-PROTEIN, FROM BK STRAIN, 20 STRUCTURES | | Descriptor: | NS3 SERINE PROTEASE, ZINC ION | | Authors: | Barbato, G, Cicero, D.O, Nardi, M.C, Steinkuhler, C, Cortese, R, De Francesco, R, Bazzo, R. | | Deposit date: | 1998-09-01 | | Release date: | 1999-06-22 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The solution structure of the N-terminal proteinase domain of the hepatitis C virus (HCV) NS3 protein provides new insights into its activation and catalytic mechanism.
J.Mol.Biol., 289, 1999
|
|
7LSE
 
 | |
7LSF
 
 | |
7LSG
 
 | |
5VIG
 
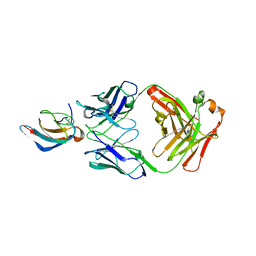 | | Crystal structure of anti-Zika antibody Z006 bound to Zika virus envelope protein DIII | | Descriptor: | CITRATE ANION, Fab heavy chain, Fab light chain, ... | | Authors: | Keeffe, J.R, West Jr, A.P, Gristick, H.B, Bjorkman, P.J. | | Deposit date: | 2017-04-16 | | Release date: | 2017-05-03 | | Last modified: | 2017-05-17 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Recurrent Potent Human Neutralizing Antibodies to Zika Virus in Brazil and Mexico.
Cell, 169, 2017
|
|
4PIK
 
 | |
4PIT
 
 | |
5VIC
 
 | | Crystal structure of anti-Zika antibody Z004 bound to DENV-1 Envelope protein DIII | | Descriptor: | Dengue 1 Envelope DIII domain, Fab heavy chain, Fab light chain | | Authors: | Keeffe, J.R, West Jr, A.P, Gristick, H.B, Bjorkman, P.J. | | Deposit date: | 2017-04-14 | | Release date: | 2017-05-03 | | Last modified: | 2017-05-17 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Recurrent Potent Human Neutralizing Antibodies to Zika Virus in Brazil and Mexico.
Cell, 169, 2017
|
|
4PIF
 
 | |
