1S4B
 
 | | Crystal structure of human thimet oligopeptidase. | | Descriptor: | Thimet oligopeptidase, ZINC ION | | Authors: | Ray, K, Hines, C.S, Coll-Rodriguez, J, Rodgers, D.W. | | Deposit date: | 2004-01-15 | | Release date: | 2004-07-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of human thimet oligopeptidase provides insight into substrate recognition, regulation, and localization
J.Biol.Chem., 279, 2004
|
|
8G83
 
 | | Structure of NAD+ consuming protein Acinetobacter baumannii TIR domain | | Descriptor: | NAD(+) hydrolase AbTIR | | Authors: | Klontz, E.H, Wang, Y, Glendening, G, Carr, J, Tsibouris, T, Buddula, S, Nallar, S, Soares, A, Snyder, G.A. | | Deposit date: | 2023-02-17 | | Release date: | 2023-10-11 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3.03 Å) | | Cite: | The structure of NAD + consuming protein Acinetobacter baumannii TIR domain shows unique kinetics and conformations.
J.Biol.Chem., 299, 2023
|
|
6OFI
 
 | | CRYSTAL STRUCTURE OF the RV144 C1-C2 SPECIFIC ANTIBODY CH55 FAB IN COMPLEX WITH HIV-1 CLADE A/E GP120 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CH55 Fab heavy chain, CH55 Fab light chain, ... | | Authors: | Tolbert, W.D, Yan, F, Van, V, Pazgier, M. | | Deposit date: | 2019-03-29 | | Release date: | 2020-06-24 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.85 Å) | | Cite: | Recognition Patterns of the C1/C2 Epitopes Involved in Fc-Mediated Response in HIV-1 Natural Infection and the RV114 Vaccine Trial.
Mbio, 11, 2020
|
|
6OED
 
 | | CRYSTAL STRUCTURE OF THE RV144 C1-C2 SPECIFIC ANTIBODY CH55 FAB | | Descriptor: | CH55 Fab heavy chain, CH55 Fab light chain | | Authors: | Yan, F, Van, V, Tolbert, W.D, Pazgier, M. | | Deposit date: | 2019-03-27 | | Release date: | 2020-07-15 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.461 Å) | | Cite: | Recognition Patterns of the C1/C2 Epitopes Involved in Fc-Mediated Response in HIV-1 Natural Infection and the RV114 Vaccine Trial.
Mbio, 11, 2020
|
|
6OEJ
 
 | |
6W4M
 
 | | CRYSTAL STRUCTURE OF THE ADCC-POTENT, WEAKLY NEUTRALIZING HIV ENV CO-RECEPTOR BINDING SITE ANTIBODY N12-I2 FAB IN COMPLEX WITH HIV-1 CLADE A/E GP120 AND M48U1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ANTI-HIV ANTIBODY N12-I2 FAB HEAVY CHAIN, ANTI-HIV ANTIBODY N12-I2 FAB LIGHT CHAIN, ... | | Authors: | Tolbert, W.D, Gohain, N, Pazgier, M. | | Deposit date: | 2020-03-11 | | Release date: | 2020-08-05 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Defining rules governing recognition and Fc-mediated effector functions to the HIV-1 co-receptor binding site.
Bmc Biol., 18, 2020
|
|
4RFE
 
 | | Crystal structure of ADCC-potent ANTI-HIV-1 Rhesus macaque antibody JR4 Fab | | Descriptor: | CHLORIDE ION, Fab heavy chain of ADCC-potent anti-HIV-1 antibody JR4, Fab light chain of ADCC-potent anti-HIV-1 antibody JR4, ... | | Authors: | Wu, X, Gohain, N, Tolbert, W.D, Pazgier, M. | | Deposit date: | 2014-09-25 | | Release date: | 2015-07-15 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Cocrystal Structures of Antibody N60-i3 and Antibody JR4 in Complex with gp120 Define More Cluster A Epitopes Involved in Effective Antibody-Dependent Effector Function against HIV-1.
J.Virol., 89, 2015
|
|
4RFO
 
 | | Crystal structure of the ADCC-Potent Antibody N60-I3 Fab in complex with HIV-1 Clade A/E gp120 and M48u1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, HIV-1 clade A/E gp120, N60-i3 Fab heavy chain, ... | | Authors: | Tolbert, W.D, Gohain, N, Pazgier, M. | | Deposit date: | 2014-09-26 | | Release date: | 2015-07-15 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Cocrystal Structures of Antibody N60-i3 and Antibody JR4 in Complex with gp120 Define More Cluster A Epitopes Involved in Effective Antibody-Dependent Effector Function against HIV-1.
J.Virol., 89, 2015
|
|
4RFN
 
 | | Crystal structure of ADCC-potent Rhesus macaque ANTIBODY JR4 in complex with HIV-1 CLADE A/E GP120 and M48 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, FAB HEAVY CHAIN OF ADCC ANTI-HIV-1 ANTIBODY JR4, FAB LIGHT CHAIN OF ADCC ANTI-HIV-1 ANTIBODY JR4, ... | | Authors: | Gohain, N, Tolbert, W.D, Pazgier, M. | | Deposit date: | 2014-09-26 | | Release date: | 2015-07-15 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.21 Å) | | Cite: | Cocrystal Structures of Antibody N60-i3 and Antibody JR4 in Complex with gp120 Define More Cluster A Epitopes Involved in Effective Antibody-Dependent Effector Function against HIV-1.
J.Virol., 89, 2015
|
|
3MPT
 
 | | Crystal structure of P38 kinase in complex with a pyrrole-2-carboxamide inhibitor | | Descriptor: | GLYCEROL, Mitogen-activated protein kinase 14, N-(furan-2-ylmethyl)-4-[(2-methylphenyl)carbonyl]-1H-pyrrole-2-carboxamide | | Authors: | Somers, D.O. | | Deposit date: | 2010-04-27 | | Release date: | 2010-07-07 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | The discovery and initial optimisation of pyrrole-2-carboxamides as inhibitors of p38alpha MAP kinase.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
2O3E
 
 | |
2O36
 
 | |
6CHF
 
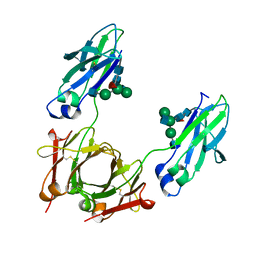 | | Crystal structure of a Fc fragment LALA mutant (L234A, L235A) of human IgG1 (crystal form 1) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Uncharacterized protein DKFZp686C11235 | | Authors: | Gohain, N, Tolbert, W.D, Pazgier, M. | | Deposit date: | 2018-02-22 | | Release date: | 2019-02-27 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Antigen-Induced Allosteric Changes in a Human IgG1 Fc Increase Low-Affinity Fc gamma Receptor Binding.
Structure, 2020
|
|
5W4L
 
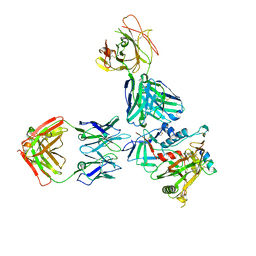 | | Crystal structure of the non-neutralizing and ADCC-potent C11-like antibody N12-i3 in complex with HIV-1 clade A/E gp120, the CD4 mimetic M48U1, and the antibody N5-i5. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Antibody N12-i3 Fab heavy chain, Antibody N12-i3 light chain, ... | | Authors: | Tolbert, W.D, Gohain, N, Pazgier, M. | | Deposit date: | 2017-06-12 | | Release date: | 2017-11-15 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.92 Å) | | Cite: | Targeting the Late Stage of HIV-1 Entry for Antibody-Dependent Cellular Cytotoxicity: Structural Basis for Env Epitopes in the C11 Region.
Structure, 25, 2017
|
|
4FZ8
 
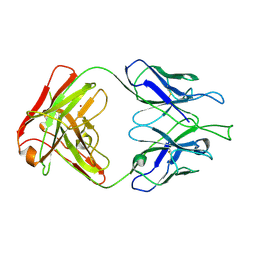 | | Crystal structure of C11 Fab, an ADCC mediating anti-HIV-1 antibody. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, FAB heavy chain of human ANTI-HIV-1 ENV ANTIBODY C11, FAB light chain of human ANTI-HIV-1 ENV ANTIBODY C11, ... | | Authors: | Wu, X, Tolbert, W.D, Pazgier, M. | | Deposit date: | 2012-07-06 | | Release date: | 2013-07-10 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.66 Å) | | Cite: | Recognition Patterns of the C1/C2 Epitopes Involved in Fc-Mediated Response in HIV-1 Natural Infection and the RV114 Vaccine Trial.
Mbio, 11, 2020
|
|
5DRZ
 
 | | Crystal structure of anti-HIV-1 antibody F240 Fab in complex with gp41 peptide | | Descriptor: | Envelope glycoprotein gp160, HIV Antibody F240 Heavy Chain, HIV Antibody F240 Light Chain, ... | | Authors: | Gohain, N, Tolbert, W.D, Pazgier, M. | | Deposit date: | 2015-09-16 | | Release date: | 2016-10-19 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | Molecular basis for epitope recognition by non-neutralizing anti-gp41 antibody F240.
Sci Rep, 6, 2016
|
|
6MG7
 
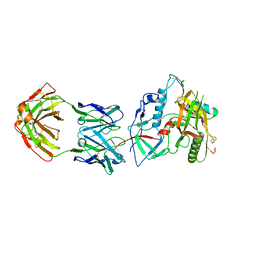 | | Crystal structure of the RV144 C1-C2 specific antibody CH54 Fab in complex with HIV-1 CLADE A/E GP120 and M48U1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CH54 Fab heavy chain, CH54 Fab light chain, ... | | Authors: | Van, V, Tolbert, W.D, Pazgier, M. | | Deposit date: | 2018-09-13 | | Release date: | 2019-09-25 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.91 Å) | | Cite: | Recognition Patterns of the C1/C2 Epitopes Involved in Fc-Mediated Response in HIV-1 Natural Infection and the RV114 Vaccine Trial.
Mbio, 11, 2020
|
|
6CJX
 
 | | Crystal structure of a Fc fragment LALA mutant (L234A, L235A) of human IgG1 (crystal form 2) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Fc fragment of human IgG1 | | Authors: | Van, V, Tolbert, W.D, Pazgier, M. | | Deposit date: | 2018-02-27 | | Release date: | 2019-03-06 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Antigen-Induced Allosteric Changes in a Human IgG1 Fc Increase Low-Affinity Fc gamma Receptor Binding.
Structure, 2020
|
|
6CJC
 
 | | CRYSTAL STRUCTURE OF A FC FRAGMENT LALA MUTANT (L234A, L235A) OF HUMAN IGG1 (CRYSTAL FORM 3) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, Uncharacterized protein DKFZp686C11235 | | Authors: | Tolbert, W.D, Van, V, Pazgier, M. | | Deposit date: | 2018-02-26 | | Release date: | 2019-03-06 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.575 Å) | | Cite: | Antigen-Induced Allosteric Changes in a Human IgG1 Fc Increase Low-Affinity Fc gamma Receptor Binding.
Structure, 28, 2020
|
|
4FXY
 
 | | Crystal structure of rat neurolysin with bound pyrazolidin inhibitor | | Descriptor: | 1-{(2S)-1-[(3R)-3-(2-chlorophenyl)-2-(2-fluorophenyl)pyrazolidin-1-yl]-1-oxopropan-2-yl}-3-[(1R,3S,5R,7R)-tricyclo[3.3.1.1~3,7~]dec-2-yl]urea, Neurolysin, mitochondrial, ... | | Authors: | Rodgers, D.W, Hines, C.S. | | Deposit date: | 2012-07-03 | | Release date: | 2013-11-13 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Allosteric inhibition of the neuropeptidase neurolysin.
J.Biol.Chem., 289, 2014
|
|
