4DM8
 
 | | Crystal structure of RARb LBD in complex with 9cis retinoic acid | | Descriptor: | Nuclear receptor coactivator 1, RETINOIC ACID, Retinoic acid receptor beta | | Authors: | Osz, J, Br livet, Y, Peluso-Iltis, C, Cura, V, Eiler, S, Ruff, M, Bourguet, W, Rochel, N, Moras, D. | | Deposit date: | 2012-02-07 | | Release date: | 2012-03-07 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for a molecular allosteric control mechanism of cofactor binding to nuclear receptors.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4DM6
 
 | | Crystal structure of RARb LBD homodimer in complex with TTNPB | | Descriptor: | 4-[(1E)-2-(5,5,8,8-TETRAMETHYL-5,6,7,8-TETRAHYDRONAPHTHALEN-2-YL)PROP-1-ENYL]BENZOIC ACID, Nuclear receptor coactivator 1, Retinoic acid receptor beta | | Authors: | Osz, J, Brelivet, Y, Peluso-Iltis, C, Cura, V, Eiler, S, Ruff, M, Bourguet, W, Rochel, N, Moras, D. | | Deposit date: | 2012-02-07 | | Release date: | 2012-03-07 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for a molecular allosteric control mechanism of cofactor binding to nuclear receptors.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4DMA
 
 | | Crystal structure of ERa LBD in complex with RU100132 | | Descriptor: | 2'-bromo-6'-(furan-3-yl)-4'-(hydroxymethyl)biphenyl-4-ol, Estrogen receptor, Nuclear receptor coactivator 1 | | Authors: | Osz, J, Brelivet, Y, Peluso-Iltis, C, Cura, V, Eiler, S, Ruff, M, Bourguet, W, Rochel, N, Moras, D. | | Deposit date: | 2012-02-07 | | Release date: | 2012-03-07 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for a molecular allosteric control mechanism of cofactor binding to nuclear receptors.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4CN2
 
 | | Crystal Structure of the Human Retinoid X Receptor DNA-Binding Domain Bound to the Human Ramp2 Response Element | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 5'-D(*AP*TP*TP*GP*AP*CP*CP*CP*TP*TP*GP*AP*AP*DC *TP*CP*AP)-3', 5'-D(*TP*GP*AP*GP*TP*TP*CP*AP*AP*GP*GP*GP*TP*DC *AP*AP*TP)-3', ... | | Authors: | McEwen, A.G, Poussin-Courmontagne, P, Osz, J, Rochel, N. | | Deposit date: | 2014-01-21 | | Release date: | 2015-02-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.069 Å) | | Cite: | Structural Basis of Natural Promoter Recognition by the Retinoid X Nuclear Receptor.
Sci.Rep., 5, 2015
|
|
4CN3
 
 | | Crystal Structure of the Human Retinoid X Receptor DNA-Binding Domain Bound to the Human Gde1SpA Response Element | | Descriptor: | 5'-D(*CP*TP*AP*GP*TP*TP*CP*AP*AP*AP*GP*TP*TP*CP *AP*CP*A)-3', 5'-D(*TP*GP*TP*GP*AP*AP*CP*TP*TP*TP*GP*AP*AP*CP *TP*AP*G)-3', RETINOIC ACID RECEPTOR RXR-ALPHA, ... | | Authors: | McEwen, A.G, Poussin-Courmontagne, P, Osz, J, Rochel, N. | | Deposit date: | 2014-01-21 | | Release date: | 2015-02-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural Basis of Natural Promoter Recognition by the Retinoid X Nuclear Receptor.
Sci.Rep., 5, 2015
|
|
4CN7
 
 | | Crystal Structure of the Human Retinoid X Receptor DNA-Binding Domain Bound to an idealized DR1 Response Element | | Descriptor: | 5'-D(*CP*TP*AP*GP*GP*TP*CP*AP*AP*AP*GP*GP*TP*CP *AP*GP)-3', 5'-D(*CP*TP*GP*AP*CP*CP*TP*TP*TP*GP*AP*CP*CP*TP *AP*GP)-3', CHLORIDE ION, ... | | Authors: | McEwen, A.G, Poussin-Courmontagne, P, Osz, J, Rochel, N. | | Deposit date: | 2014-01-21 | | Release date: | 2015-02-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Structural Basis of Natural Promoter Recognition by the Retinoid X Nuclear Receptor.
Sci.Rep., 5, 2015
|
|
4CN5
 
 | | Crystal Structure of the Human Retinoid X Receptor DNA-Binding Domain Bound to the Human Nr1d1 Response Element | | Descriptor: | 5'-D(*AP*TP*TP*GP*AP*AP*CP*TP*CP*TP*GP*AP*CP*CP *CP*CP*AP)-3', 5'-D(*TP*GP*GP*GP*GP*TP*CP*AP*GP*AP*GP*TP*TP*CP *AP*AP*TP)-3', CHLORIDE ION, ... | | Authors: | McEwen, A.G, Poussin-Courmontagne, P, Osz, J, Rochel, N. | | Deposit date: | 2014-01-21 | | Release date: | 2015-02-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Basis of Natural Promoter Recognition by the Retinoid X Nuclear Receptor.
Sci.Rep., 5, 2015
|
|
6FBR
 
 | | Crystal Structure of the Human Retinoid X Receptor DNA-Binding Domain Bound to the Human MEp DR1 Response Element, pH 4.2 | | Descriptor: | DI(HYDROXYETHYL)ETHER, DNA (5'-D(*CP*TP*GP*GP*GP*TP*CP*AP*AP*AP*GP*TP*TP*CP*AP*TP*C)-3'), DNA (5'-D(*GP*AP*TP*GP*AP*AP*CP*TP*TP*TP*GP*AP*CP*CP*CP*AP*G)-3'), ... | | Authors: | McEwen, A.G, Poussin-Courmontagne, P, Osz, J, Rochel, N. | | Deposit date: | 2017-12-19 | | Release date: | 2018-12-05 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Modulation of RXR-DNA complex assembly by DNA context.
Mol. Cell. Endocrinol., 481, 2019
|
|
6FBQ
 
 | | Crystal Structure of the Human Retinoid X Receptor DNA-Binding Domain Bound to the Human MEp DR1 Response Element, pH 7.0 | | Descriptor: | 3[N-MORPHOLINO]PROPANE SULFONIC ACID, DNA (5'-D(*CP*TP*GP*GP*GP*TP*CP*AP*AP*AP*GP*TP*TP*CP*AP*TP*C)-3'), DNA (5'-D(*GP*AP*TP*GP*AP*AP*CP*TP*TP*TP*GP*AP*CP*CP*CP*AP*G)-3'), ... | | Authors: | McEwen, A.G, Poussin-Courmontagne, P, Osz, J, Rochel, N. | | Deposit date: | 2017-12-19 | | Release date: | 2018-12-05 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Modulation of RXR-DNA complex assembly by DNA context.
Mol. Cell. Endocrinol., 481, 2019
|
|
6ZYU
 
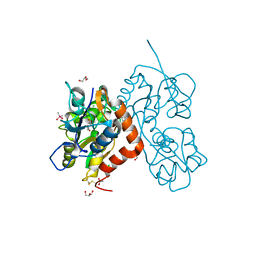 | | Structure of the GluA2 ligand-binding domain (L483Y-N754S) in complex with glutamate and BPAM549 | | Descriptor: | (4~{R})-4-cyclopropyl-7-fluoranyl-3,4-dihydro-2~{H}-thiochromene 1,1-dioxide, (4~{S})-4-cyclopropyl-7-fluoranyl-3,4-dihydro-2~{H}-thiochromene 1,1-dioxide, ACETATE ION, ... | | Authors: | Dorosz, J, Christensen, K.M, Kastrup, J.S. | | Deposit date: | 2020-08-03 | | Release date: | 2021-08-11 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Development of Thiochroman Dioxide Analogues of Benzothiadiazine Dioxides as New Positive Allosteric Modulators of alpha-Amino-3-hydroxy-5-methyl-4-isoxazolepropionic Acid (AMPA) Receptors.
Acs Chem Neurosci, 12, 2021
|
|
8QEZ
 
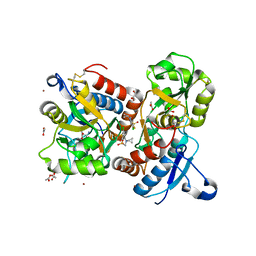 | | Crystal structure of the AMPA receptor GluA2-L504Y-N775S ligand binding domain in complex with L-glutamate and positive allosteric modulator BPAM395 at 1.55A resolution | | Descriptor: | 6-chloranyl-4-cyclopropyl-2,3-dihydrothieno[3,2-e][1,2,4]thiadiazine 1,1-dioxide, ACETATE ION, CACODYLATE ION, ... | | Authors: | Dorosz, J, Laulumaa, S, Frydenvang, K, Kastrup, J.S. | | Deposit date: | 2023-09-01 | | Release date: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Exploring thienothiadiazine dioxides as isosteric analogues of benzo- and pyridothiadiazine dioxides in the search of new AMPA and kainate receptor positive allosteric modulators.
Eur.J.Med.Chem., 264, 2023
|
|
6SQC
 
 | | Crystal structure of complex between nuclear coactivator binding domain of CBP and [1040-1086]ACTR containing alpha-methylated Leu1055 and Leu1076 | | Descriptor: | 1,2-ETHANEDIOL, Maltose/maltodextrin-binding periplasmic protein,CREB-binding protein, Nuclear receptor coactivator 3, ... | | Authors: | Bauer, V, Schmidtgall, B, Gogl, G, Dolenc, j, Osz, J, Kostmann, C, Mitschler, A, Cousido-Siah, A, Rochel, N, Trave, G, Kieffer, B, Torbeev, V. | | Deposit date: | 2019-09-03 | | Release date: | 2020-09-30 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Conformational editing of intrinsically disordered protein by alpha-methylation.
Chem Sci, 12, 2020
|
|
9C5E
 
 | | Covalent Complex Between Parkin Catalytic (Rcat) Domain and Ubiquitin | | Descriptor: | 1.7.6 3-bromanylpropan-1-amine, E3 ubiquitin-protein ligase parkin, Ubiquitin, ... | | Authors: | Connelly, E.M, Rintala-Dempsey, A.C, Gundogdu, M, Freeman, E.A, Koszela, J, Aguirre, J.D, Zhu, G, Kamarainen, O, Tadayon, R, Walden, H, Shaw, G.S. | | Deposit date: | 2024-06-06 | | Release date: | 2024-08-14 | | Method: | SOLUTION NMR | | Cite: | Capturing the catalytic intermediates of parkin ubiquitination.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
6XWG
 
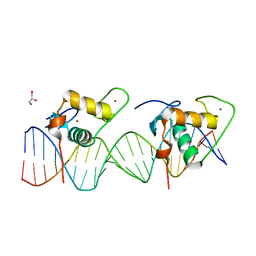 | | Crystal Structure of the Human RXR/RAR DNA-Binding Domain Heterodimer Bound to the Human RARb2 DR5 Response Element | | Descriptor: | CHLORIDE ION, GLYCEROL, RARb2 DR5 Response Element, ... | | Authors: | McEwen, A.G, Poussin-Courmontagne, P, Peluso-Iltis, C, Rochel, N. | | Deposit date: | 2020-01-23 | | Release date: | 2020-09-09 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis for DNA recognition and allosteric control of the retinoic acid receptors RAR-RXR.
Nucleic Acids Res., 48, 2020
|
|
6XWH
 
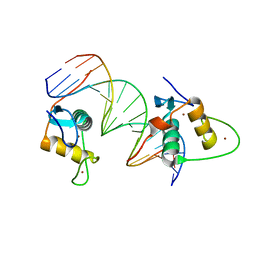 | | Crystal Structure of the Human RXR DNA-Binding Domain Homodimer Bound to the Human Hoxb13 DR0 Response Element | | Descriptor: | Hoxb13 DR0 Response Element, 3'-5' strand, 5'-3' strand, ... | | Authors: | McEwen, A.G, Poussin-Courmontagne, P, Peluso-Iltis, C, Rochel, N. | | Deposit date: | 2020-01-23 | | Release date: | 2020-09-09 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis for DNA recognition and allosteric control of the retinoic acid receptors RAR-RXR.
Nucleic Acids Res., 48, 2020
|
|
6XZV
 
 | | Structure of zVDR LBD-Calcitriol in complex with chimera 18 | | Descriptor: | 5-{2-[1-(5-HYDROXY-1,5-DIMETHYL-HEXYL)-7A-METHYL-OCTAHYDRO-INDEN-4-YLIDENE]-ETHYLIDENE}-4-METHYLENE-CYCLOHEXANE-1,3-DIOL, URA-UIA-URL-URY-URV-UZN-LYS, Vitamin D3 receptor A | | Authors: | Buratto, J, Belorusova, A.Y, Rochel, N, Guichard, G. | | Deposit date: | 2020-02-05 | | Release date: | 2021-02-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Basis for alpha-Helix Mimicry and Inhibition of Protein-Protein Interactions with Oligourea Foldamers.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
3A9E
 
 | | Crystal structure of a mixed agonist-bound RAR-alpha and antagonist-bound RXR-alpha heterodimer ligand binding domains | | Descriptor: | (2E,4E,6Z)-3-methyl-7-(5,5,8,8-tetramethyl-3-propoxy-5,6,7,8-tetrahydronaphthalen-2-yl)octa-2,4,6-trienoic acid, 13-mer (LXXLL motif) from Nuclear receptor coactivator 2, RETINOIC ACID, ... | | Authors: | Sato, Y, Duclaud, S, Peluso-Iltis, C, Poussin, P, Moras, D, Rochel, N, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2009-10-24 | | Release date: | 2010-10-06 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | The Phantom Effect of the Rexinoid LG100754: structural and functional insights
Plos One, 5, 2010
|
|
6XZJ
 
 | | Structure of zVDR LBD-Calcitriol in complex with chimera 12 | | Descriptor: | 5-{2-[1-(5-HYDROXY-1,5-DIMETHYL-HEXYL)-7A-METHYL-OCTAHYDRO-INDEN-4-YLIDENE]-ETHYLIDENE}-4-METHYLENE-CYCLOHEXANE-1,3-DIOL, ACETATE ION, ARG-HIS-LYS-ILE-LEU-URR-UIL-URL-GLN, ... | | Authors: | Buratto, J, Belorusova, A.Y, Rochel, N, Guichard, G. | | Deposit date: | 2020-02-04 | | Release date: | 2021-02-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Basis for alpha-Helix Mimicry and Inhibition of Protein-Protein Interactions with Oligourea Foldamers.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
6XZH
 
 | | Structure of zVDR LBD-Calcitriol in complex with chimera 10 | | Descriptor: | 5-{2-[1-(5-HYDROXY-1,5-DIMETHYL-HEXYL)-7A-METHYL-OCTAHYDRO-INDEN-4-YLIDENE]-ETHYLIDENE}-4-METHYLENE-CYCLOHEXANE-1,3-DIOL, ARG-HIS-LYS-ILE-URL-URK-URL-LEU-GLN, Vitamin D3 receptor A | | Authors: | Buratto, J, Belorusova, A.Y, Rochel, N, Guichard, G. | | Deposit date: | 2020-02-04 | | Release date: | 2021-02-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.372 Å) | | Cite: | Structural Basis for alpha-Helix Mimicry and Inhibition of Protein-Protein Interactions with Oligourea Foldamers.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
6XZI
 
 | | Structure of zVDR LBD-calcitriol in complex with chimera 11 | | Descriptor: | 5-{2-[1-(5-HYDROXY-1,5-DIMETHYL-HEXYL)-7A-METHYL-OCTAHYDRO-INDEN-4-YLIDENE]-ETHYLIDENE}-4-METHYLENE-CYCLOHEXANE-1,3-DIOL, ACETATE ION, ARG-HIS-LYS-ILE-LEU-URK-UIL-URL, ... | | Authors: | Buratto, J, Belorusova, A.Y, Rochel, N, Guichard, G. | | Deposit date: | 2020-02-04 | | Release date: | 2021-02-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Basis for alpha-Helix Mimicry and Inhibition of Protein-Protein Interactions with Oligourea Foldamers.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
6XZK
 
 | | Structure of zVDR LBD-Calcitriol in complex with chimera 13 | | Descriptor: | 5-{2-[1-(5-HYDROXY-1,5-DIMETHYL-HEXYL)-7A-METHYL-OCTAHYDRO-INDEN-4-YLIDENE]-ETHYLIDENE}-4-METHYLENE-CYCLOHEXANE-1,3-DIOL, ACETATE ION, GLU-ASN-ALA-UIA-URL-URY-URV-UZN-LYS, ... | | Authors: | Buratto, J, Belorusova, A.Y, Rochel, N, Guichard, G. | | Deposit date: | 2020-02-04 | | Release date: | 2021-02-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Basis for alpha-Helix Mimicry and Inhibition of Protein-Protein Interactions with Oligourea Foldamers.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
9EZ1
 
 | | Vitamin D receptor in complex with 1,4a,25-trihydroxyvitamin D3 | | Descriptor: | 1,4a,25-trihydroxyvitamin D3, ACETATE ION, Nuclear receptor coactivator 2, ... | | Authors: | Rochel, N. | | Deposit date: | 2024-04-10 | | Release date: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | 4-Hydroxy-1 alpha ,25-Dihydroxyvitamin D 3 : Synthesis and Structure-Function Study.
Biomolecules, 14, 2024
|
|
9EZ2
 
 | | Vitamin D receptor complex with 1,4b,25-trihydroxyvitamin D3 | | Descriptor: | 1,4b,25-trihydroxyvitamin D3, ACETATE ION, Nuclear receptor coactivator 2, ... | | Authors: | Rochel, N. | | Deposit date: | 2024-04-10 | | Release date: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | 4-Hydroxy-1 alpha ,25-Dihydroxyvitamin D 3 : Synthesis and Structure-Function Study.
Biomolecules, 14, 2024
|
|
7QP4
 
 | | Complex of a Gemini-cholesterol analogue with Retinoid-related Orphan Receptor gamma | | Descriptor: | (3~{S},8~{S},9~{S},10~{R},13~{R},14~{S},17~{R})-17-[(6~{R})-2,10-dimethyl-2-oxidanyl-undecan-6-yl]-10,13-dimethyl-2,3,4,7,8,9,11,12,14,15,16,17-dodecahydro-1~{H}-cyclopenta[a]phenanthren-3-ol, ACETATE ION, HIS-VAL-GLU-ARG-LEU-GLN-ILE-PHE-GLN-HIS-LEU-HIS-PRO-ILE-VAL, ... | | Authors: | Rochel, N. | | Deposit date: | 2022-01-02 | | Release date: | 2022-12-14 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Development of novel Gemini-cholesterol analogues for retinoid-related orphan receptors
Org Chem Front, 9, 2022
|
|
7QPI
 
 | | Structure of lamprey VDR in complex with 1,25D3 | | Descriptor: | 5-{2-[1-(5-HYDROXY-1,5-DIMETHYL-HEXYL)-7A-METHYL-OCTAHYDRO-INDEN-4-YLIDENE]-ETHYLIDENE}-4-METHYLENE-CYCLOHEXANE-1,3-DIOL, MAGNESIUM ION, Nuclear receptor coactivator 1, ... | | Authors: | Rochel, N. | | Deposit date: | 2022-01-04 | | Release date: | 2022-12-14 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Advances in Vitamin D Receptor Function and Evolution Based on the 3D Structure of the Lamprey Ligand-Binding Domain.
J.Med.Chem., 65, 2022
|
|
