2V5L
 
 | | Structures of the Open and Closed State of Trypanosomal Triosephosphate Isomerase: as Observed in a New Crystal Form: Implications for the Reaction Mechanism | | Descriptor: | SULFATE ION, TRIOSEPHOSPHATE ISOMERASE | | Authors: | Noble, M.E.M, Zeelen, J.P, Wierenga, R.K. | | Deposit date: | 2007-07-06 | | Release date: | 2007-07-31 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structures of the Open and Closed State of Trypanosomal Triosephosphate Isomerase: As Observed in a New Crystal Form: Implications for the Reaction Mechanism
Proteins: Struct.,Funct., Genet., 16, 1993
|
|
6TIM
 
 | |
1VIN
 
 | |
8ROZ
 
 | |
6SG4
 
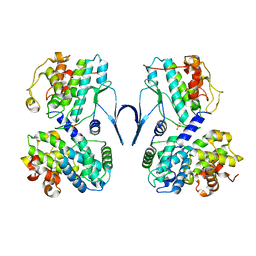 | | Structure of CDK2/cyclin A M246Q, S247EN | | Descriptor: | Cyclin-A2, Cyclin-dependent kinase 2 | | Authors: | Salamina, M, Basle, A, Massa, B, Noble, M.E.M, Endicott, J.A. | | Deposit date: | 2019-08-02 | | Release date: | 2021-01-27 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Discriminative SKP2 Interactions with CDK-Cyclin Complexes Support a Cyclin A-Specific Role in p27KIP1 Degradation.
J.Mol.Biol., 433, 2021
|
|
2CCH
 
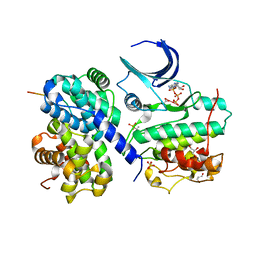 | | The crystal structure of CDK2 cyclin A in complex with a substrate peptide derived from CDC modified with a gamma-linked ATP analogue | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CELL DIVISION CONTROL PROTEIN 6 HOMOLOG, CELL DIVISION PROTEIN KINASE 2, ... | | Authors: | Cheng, K.Y, Noble, M.E.M, Skamnaki, V, Brown, N.R, Lowe, E.D, Kontogiannis, L, Shen, K, Cole, P.A, Siligardi, G, Johnson, L.N. | | Deposit date: | 2006-01-16 | | Release date: | 2006-05-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The Role of the Phospho-Cdk2/Cyclin a Recruitment Site in Substrate Recognition
J.Biol.Chem., 281, 2006
|
|
7OZT
 
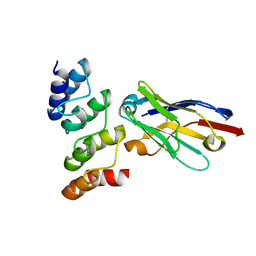 | | Nanobodies restore stability to cancer-associated mutants of tumor suppressor protein p16INK4a | | Descriptor: | Camelid nanobody NB09, Cyclin-dependent kinase inhibitor 2A | | Authors: | Pastok, M.W, Burbidge, O, Itzhaki, L, Endicott, J.A, Noble, M.E.M. | | Deposit date: | 2021-06-28 | | Release date: | 2022-07-13 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Nanobodies restore stability to cancer-associated mutants of tumor suppressor protein p16INK4a
To Be Published
|
|
7QUM
 
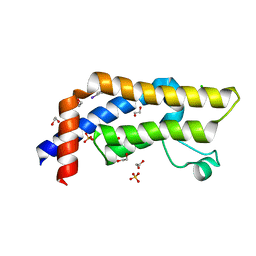 | | ATAD2 in complex with FragLite2 | | Descriptor: | 1,2-ETHANEDIOL, 4-IODOPYRAZOLE, ATPase family AAA domain-containing protein 2, ... | | Authors: | Turberville, S, Martin, M.P, Hope, I, Noble, M.E.M. | | Deposit date: | 2022-01-18 | | Release date: | 2023-03-01 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Mapping Ligand Interactions of Bromodomains BRD4 and ATAD2 with FragLites and PepLites─Halogenated Probes of Druglike and Peptide-like Molecular Interactions.
J.Med.Chem., 65, 2022
|
|
7QZM
 
 | | ATAD2 in complex with FragLite28 | | Descriptor: | 1,2-ETHANEDIOL, 4-bromo-1-(2-hydroxyethyl)pyridin-2(1H)-one, ATPase family AAA domain-containing protein 2, ... | | Authors: | Turberville, S, Martin, M.P, Hope, I, Noble, M.E.M. | | Deposit date: | 2022-01-31 | | Release date: | 2023-03-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Mapping Ligand Interactions of Bromodomains BRD4 and ATAD2 with FragLites and PepLites─Halogenated Probes of Druglike and Peptide-like Molecular Interactions.
J.Med.Chem., 65, 2022
|
|
7QZY
 
 | | ATAD2 in complex with FragLite29 | | Descriptor: | 1,2-ETHANEDIOL, 4-bromanyl-1-(2-methoxyethyl)pyridin-2-one, ATPase family AAA domain-containing protein 2, ... | | Authors: | Turberville, S, Martin, M.P, Hope, I, Noble, M.E.M. | | Deposit date: | 2022-02-01 | | Release date: | 2023-03-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Mapping Ligand Interactions of Bromodomains BRD4 and ATAD2 with FragLites and PepLites─Halogenated Probes of Druglike and Peptide-like Molecular Interactions.
J.Med.Chem., 65, 2022
|
|
7QYK
 
 | | ATAD2 in complex with FragLite18 | | Descriptor: | (4-bromanylpyridin-2-yl)methanol, 1,2-ETHANEDIOL, ATPase family AAA domain-containing protein 2, ... | | Authors: | Turberville, S, Martin, M.P, Hope, I, Noble, M.E.M. | | Deposit date: | 2022-01-28 | | Release date: | 2023-03-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Mapping Ligand Interactions of Bromodomains BRD4 and ATAD2 with FragLites and PepLites─Halogenated Probes of Druglike and Peptide-like Molecular Interactions.
J.Med.Chem., 65, 2022
|
|
1OW6
 
 | | Paxillin LD4 motif bound to the Focal Adhesion Targeting (FAT) domain of the Focal Adhesion Kinase | | Descriptor: | Focal adhesion kinase 1, Paxillin | | Authors: | Hoellerer, M.K, Noble, M.E.M, Labesse, G, Campbell, I.D, Werner, J.M, Arold, S.T. | | Deposit date: | 2003-03-28 | | Release date: | 2003-10-21 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Molecular Recognition of Paxillin LD motifs
by the Focal Adhesion Targeting Domain
Structure, 11, 2003
|
|
5LRQ
 
 | | BRD4 in complex with ERK5 inhibitor XMD8-92 | | Descriptor: | 2-{[2-ethoxy-4-(4-hydroxypiperidin-1-yl)phenyl]amino}-5,11-dimethyl-5,11-dihydro-6H-pyrimido[4,5-b][1,4]benzodiazepin-6-one, Bromodomain-containing protein 4 | | Authors: | Martin, M.P, Noble, M.E.M. | | Deposit date: | 2016-08-19 | | Release date: | 2017-08-30 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Identification of a novel orally bioavailable ERK5 inhibitor with selectivity over p38 alpha and BRD4.
Eur.J.Med.Chem., 178, 2019
|
|
2VF2
 
 | | X-ray crystal structure of HsaD from Mycobacterium tuberculosis | | Descriptor: | 2-HYDROXY-6-OXO-6-PHENYLHEXA-2,4-DIENOATE HYDROLASE BPHD, GLYCEROL, SULFATE ION | | Authors: | Lack, N, Lowe, E.D, Liu, J, Eltis, L.D, Noble, M.E.M, Sim, E, Westwood, I.M. | | Deposit date: | 2007-10-29 | | Release date: | 2007-11-06 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structure of Hsad, a Steroid-Degrading Hydrolase, from Mycobacterium Tuberculosis.
Acta Crystallogr.,Sect.F, 64, 2008
|
|
2VFC
 
 | | The structure of Mycobacterium marinum arylamine N-acetyltransferase in complex with CoA | | Descriptor: | ARYLAMINE N-ACETYLTRANSFERASE, COENZYME A | | Authors: | Fullam, E, Westwood, I.M, Anderton, M.C, Lowe, E.D, Sim, E, Noble, M.E.M. | | Deposit date: | 2007-11-02 | | Release date: | 2007-12-18 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Divergence of Cofactor Recognition Across Evolution: Coenzyme a Binding in a Prokaryotic Arylamine N-Acetyltransferase.
J.Mol.Biol., 375, 2008
|
|
2V9C
 
 | | X-ray Crystallographic Structure of a Pseudomonas aeruginosa Azoreductase in Complex with Methyl Red. | | Descriptor: | 2-(4-DIMETHYLAMINOPHENYL)DIAZENYLBENZOIC ACID, FLAVIN MONONUCLEOTIDE, FMN-DEPENDENT NADH-AZOREDUCTASE 1, ... | | Authors: | Wang, C.-J, Hagemeier, C, Rahman, N, Lowe, E.D, Noble, M.E.M, Coughtrie, M, Sim, E, Westwood, I.M. | | Deposit date: | 2007-08-23 | | Release date: | 2007-11-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Molecular Cloning, Characterisation and Ligand- Bound Structure of an Azoreductase from Pseudomonas Aeruginosa
J.Mol.Biol., 373, 2007
|
|
2VFB
 
 | | The structure of Mycobacterium marinum arylamine N-acetyltransferase | | Descriptor: | ARYLAMINE N-ACETYLTRANSFERASE | | Authors: | Fullam, E, Westwood, I.M, Anderton, M.C, Lowe, E.D, Sim, E, Noble, M.E.M. | | Deposit date: | 2007-11-02 | | Release date: | 2007-12-18 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Divergence of Cofactor Recognition Across Evolution: Coenzyme a Binding in a Prokaryotic Arylamine N-Acetyltransferase.
J.Mol.Biol., 375, 2008
|
|
8BYL
 
 | | Cryo-EM structure of SKP1-SKP2-CKS1 from the SCFSKP2 E3 ligase complex | | Descriptor: | Cyclin-dependent kinase inhibitor 1B, Cyclin-dependent kinases regulatory subunit 1, S-phase kinase-associated protein 1, ... | | Authors: | Rowland, R.J, Salamina, M, Endicott, J.A, Noble, M.E.M. | | Deposit date: | 2022-12-13 | | Release date: | 2023-06-28 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Cryo-EM structure of SKP1-SKP2-CKS1 in complex with CDK2-cyclin A-p27KIP1.
Sci Rep, 13, 2023
|
|
2WMA
 
 | |
2WMB
 
 | | Structural and thermodynamic consequences of cyclization of peptide ligands for the recruitment site of cyclin A | | Descriptor: | CELL DIVISION PROTEIN KINASE 2, CYCLIN-A2, LINEAR RKLFD, ... | | Authors: | Robertson, G.F, Endicott, J.A, Noble, M.E.M, McDonnell, J.M. | | Deposit date: | 2009-06-30 | | Release date: | 2010-09-01 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural and Thermodynamic Consequences of Cyclization of Peptide Ligands for the Recruitment Site of Cyclin A
To be Published
|
|
2VZC
 
 | | Crystal structure of the C-terminal calponin homology domain of alpha parvin | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ... | | Authors: | Lorenz, S, Vakonakis, I, Lowe, E.D, Campbell, I.D, Noble, M.E.M, Hoellerer, M.K. | | Deposit date: | 2008-07-31 | | Release date: | 2008-10-28 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Structural Analysis of the Interactions between Paxillin Ld Motifs and Alpha-Parvin
Structure, 16, 2008
|
|
1QL6
 
 | | THE CATALYTIC MECHANISM OF PHOSPHORYLASE KINASE PROBED BY MUTATIONAL STUDIES | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MANGANESE (II) ION, PHOSPHORYLASE KINASE, ... | | Authors: | Skamnaki, V.T, Owen, D.J, Noble, M.E.M, Lowe, E.D, Oikonomakos, N.G, Johnson, L.N. | | Deposit date: | 1999-08-24 | | Release date: | 1999-12-14 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Catalytic Mechanism of Phosphorylase Kinase Probed by Mutational Studies.
Biochemistry, 38, 1999
|
|
2VZD
 
 | | Crystal structure of the C-terminal calponin homology domain of alpha parvin in complex with paxillin LD1 motif | | Descriptor: | 1,2-ETHANEDIOL, ALPHA-PARVIN, GLYCEROL, ... | | Authors: | Lorenz, S, Vakonakis, I, Lowe, E.D, Campbell, I.D, Noble, M.E.M, Hoellerer, M.K. | | Deposit date: | 2008-07-31 | | Release date: | 2008-10-28 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Analysis of the Interactions between Paxillin Ld Motifs and Alpha-Parvin
Structure, 16, 2008
|
|
1H24
 
 | | CDK2/CyclinA in complex with a 9 residue recruitment peptide from E2F | | Descriptor: | CELL DIVISION PROTEIN KINASE 2, CYCLIN A2, TRANSCRIPTION FACTOR E2F1 | | Authors: | Tews, I, Cheng, K.Y, Lowe, E.D, Noble, M.E.M, Brown, N.R, Gul, S, Gamblin, S, Johnson, L.N. | | Deposit date: | 2002-07-31 | | Release date: | 2003-02-01 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Specificity Determinants of Recruitment Peptides Bound to Phospho-Cdk2/Cyclin A
Biochemistry, 41, 2002
|
|
1H28
 
 | | CDK2/CyclinA in complex with an 11-residue recruitment peptide from p107 | | Descriptor: | CELL DIVISION PROTEIN KINASE 2, CYCLIN A2, RETINOBLASTOMA-LIKE PROTEIN 1 | | Authors: | Tews, I, Cheng, K.Y, Lowe, E.D, Noble, M.E.M, Brown, N.R, Gul, S, Gamblin, S, Johnson, L.N. | | Deposit date: | 2002-07-31 | | Release date: | 2003-02-01 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Specificity Determinants of Recruitment Peptides Bound to Phospho-Cdk2/Cyclin A
Biochemistry, 41, 2002
|
|
