1AP0
 
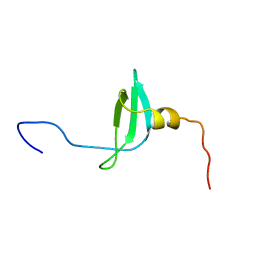 | | STRUCTURE OF THE CHROMATIN BINDING (CHROMO) DOMAIN FROM MOUSE MODIFIER PROTEIN 1, NMR, 26 STRUCTURES | | Descriptor: | MODIFIER PROTEIN 1 | | Authors: | Ball, L.J, Murzina, N.V, Broadhurst, R.W, Raine, A.R.C, Archer, S.J, Stott, F.J, Murzin, A.G, Singh, P.B, Domaille, P.J, Laue, E.D. | | Deposit date: | 1997-07-22 | | Release date: | 1998-07-22 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure of the chromatin binding (chromo) domain from mouse modifier protein 1.
EMBO J., 16, 1997
|
|
2BUD
 
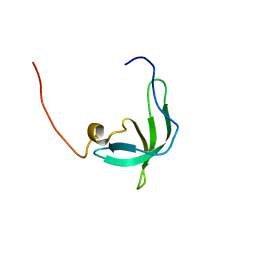 | | The solution structure of the chromo barrel domain from the males- absent on the first (MOF) protein | | Descriptor: | MALES-ABSENT ON THE FIRST PROTEIN | | Authors: | Nielsen, P.R, Nietlispach, D, Buscaino, A, Warner, R.J, Akhtar, A, Murzin, A.G, Murzina, N.V, Laue, E.D. | | Deposit date: | 2005-06-09 | | Release date: | 2005-06-24 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure of the Chromo Barrel Domain from the Mof Acetyltransferase
J.Biol.Chem., 280, 2005
|
|
9GG1
 
 | | P301T type I tau filaments from human brain | | Descriptor: | Isoform Tau-D of Microtubule-associated protein tau | | Authors: | Schweighauser, M, Shi, Y, Murzin, A.G, Garringer, H.J, Vidal, R, Murrell, J.R, Erro, M.E, Seelaar, H, Ferrer, I, van Swieten, J.C, Ghetti, B, Scheres, S.H.W, Goedert, M. | | Deposit date: | 2024-08-12 | | Release date: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (2.26 Å) | | Cite: | Novel tau filament folds in individuals with MAPT mutations P301L and P301T.
Biorxiv, 2024
|
|
9GG0
 
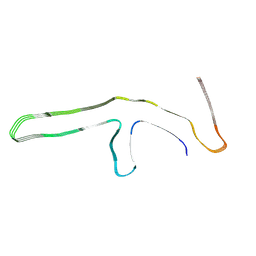 | | P301L tau filaments from human brain | | Descriptor: | Isoform Tau-D of Microtubule-associated protein tau | | Authors: | Schweighauser, M, Shi, Y, Murzin, A.G, Garringer, H.J, Vidal, R, Murrell, J.R, Erro, M.E, Seelaar, H, Ferrer, I, van Swieten, J.C, Ghetti, B, Scheres, S.H.W, Goedert, M. | | Deposit date: | 2024-08-12 | | Release date: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (2.81 Å) | | Cite: | Novel tau filament folds in individuals with MAPT mutations P301L and P301T.
Biorxiv, 2024
|
|
9GG6
 
 | | P301T type II tau filaments from human brain | | Descriptor: | Isoform Tau-F of Microtubule-associated protein tau | | Authors: | Schweighauser, M, Shi, Y, Murzin, A.G, Garringer, H.J, Vidal, R, Murrell, J.R, Erro, M.E, Seelaar, H, Ferrer, I, van Swieten, J.C, Ghetti, B, Scheres, S.H.W, Goedert, M. | | Deposit date: | 2024-08-13 | | Release date: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (3.36 Å) | | Cite: | Novel tau filament folds in individuals with MAPT mutations P301L and P301T.
Biorxiv, 2024
|
|
6VPS
 
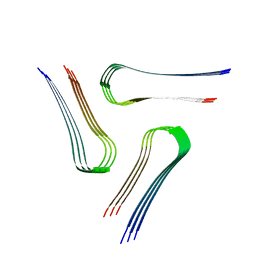 | | Cryo-EM structure of the amyloid core of Drosophila Orb2 isolated from head | | Descriptor: | Translational regulator orb2 | | Authors: | Hervas, R, Rau, M.J, Park, Y, Zhang, W, Murzin, A.G, Fitzpatrick, J.A.J, Scheres, S.H.W, Si, K. | | Deposit date: | 2020-02-04 | | Release date: | 2020-03-18 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Cryo-EM structure of a neuronal functional amyloid implicated in memory persistence in Drosophila
Science, 367, 2020
|
|
1Z0R
 
 | | Solution Structure of the N-terminal DNA Recognition Domain of the Bacillus subtilis Transcription-State Regulator AbrB | | Descriptor: | Transition state regulatory protein abrB | | Authors: | Bobay, B.G, Andreeva, A, Mueller, G.A, Cavanagh, J, Murzin, A.G. | | Deposit date: | 2005-03-02 | | Release date: | 2005-03-15 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Revised structure of the AbrB N-terminal domain unifies a diverse superfamily of putative DNA-binding proteins.
Febs Lett., 579, 2005
|
|
6NWQ
 
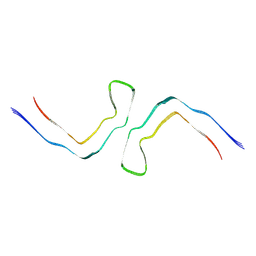 | | Chronic traumatic encephalopathy Type II Tau filament | | Descriptor: | Microtubule-associated protein tau | | Authors: | Falcon, B, Zivanov, J, Zhang, W, Murzin, A.G, Garringer, H.J, Vidal, R, Crowther, R.A, Newell, K.L, Ghetti, B, Goedert, M, Scheres, H.W. | | Deposit date: | 2019-02-07 | | Release date: | 2019-03-27 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Novel tau filament fold in chronic traumatic encephalopathy encloses hydrophobic molecules.
Nature, 568, 2019
|
|
6NWP
 
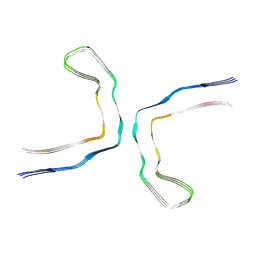 | | Chronic traumatic encephalopathy Type I Tau filament | | Descriptor: | Microtubule-associated protein tau | | Authors: | Falcon, B, Zivanov, J, Zhang, W, Murzin, A.G, Garringer, H.J, Vidal, R, Crowther, R.A, Newell, K.L, Ghetti, B, Goedert, M, Scheres, H.W. | | Deposit date: | 2019-02-07 | | Release date: | 2019-03-27 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (2.3 Å) | | Cite: | Novel tau filament fold in chronic traumatic encephalopathy encloses hydrophobic molecules.
Nature, 568, 2019
|
|
5O3L
 
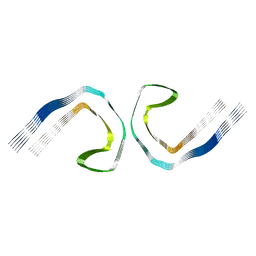 | | Paired helical filament in Alzheimer's disease brain | | Descriptor: | Microtubule-associated protein tau | | Authors: | Fitzpatrick, A.W.P, Falcon, B, He, S, Murzin, A.G, Murshudov, G, Garringer, H.G, Crowther, R.A, Ghetti, B, Goedert, M, Scheres, S.H.W. | | Deposit date: | 2017-05-24 | | Release date: | 2017-07-26 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Cryo-EM structures of tau filaments from Alzheimer's disease.
Nature, 547, 2017
|
|
5O3O
 
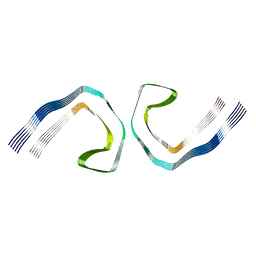 | | Pronase-treated paired helical filament in Alzheimer's disease brain | | Descriptor: | Microtubule-associated protein tau | | Authors: | Fitzpatrick, A.W.P, Falcon, B, He, S, Murzin, A.G, Murshudov, G, Garringer, H.G, Crowther, R.A, Ghetti, B, Goedert, M, Scheres, S.H.W. | | Deposit date: | 2017-05-24 | | Release date: | 2017-07-26 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Cryo-EM structures of tau filaments from Alzheimer's disease.
Nature, 547, 2017
|
|
5O3T
 
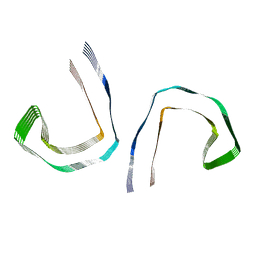 | | Straight filament in Alzheimer's disease brain | | Descriptor: | Microtubule-associated protein tau | | Authors: | Fitzpatrick, A.W.P, Falcon, B, He, S, Murzin, A.G, Murshudov, G, Garringer, H.G, Crowther, R.A, Ghetti, B, Goedert, M, Scheres, S.H.W. | | Deposit date: | 2017-05-24 | | Release date: | 2017-07-26 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Cryo-EM structures of tau filaments from Alzheimer's disease.
Nature, 547, 2017
|
|
6HRE
 
 | | Paired helical filament from sporadic Alzheimer's disease brain | | Descriptor: | Microtubule-associated protein tau | | Authors: | Falcon, B, Zhang, W, Schweighauser, M, Murzin, A.G, Vidal, R, Garringer, H.J, Ghetti, B, Scheres, S.H.W, Goedert, M. | | Deposit date: | 2018-09-26 | | Release date: | 2018-10-10 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Tau filaments from multiple cases of sporadic and inherited Alzheimer's disease adopt a common fold.
Acta Neuropathol., 136, 2018
|
|
6HRF
 
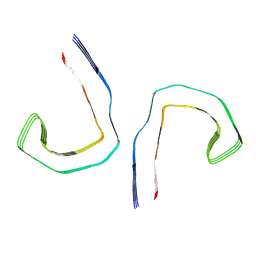 | | Straight filament from sporadic Alzheimer's disease brain | | Descriptor: | Microtubule-associated protein tau | | Authors: | Falcon, B, Zhang, W, Schweighauser, M, Murzin, A.G, Vidal, R, Garringer, H.J, Ghetti, B, Scheres, S.H.W, Goedert, M. | | Deposit date: | 2018-09-26 | | Release date: | 2018-10-10 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Tau filaments from multiple cases of sporadic and inherited Alzheimer's disease adopt a common fold.
Acta Neuropathol., 136, 2018
|
|
6GX5
 
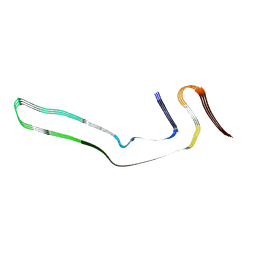 | | Narrow Pick Filament from Pick's disease brain | | Descriptor: | Microtubule-associated protein tau | | Authors: | Falcon, B, Zhang, W, Murzin, A.G, Murshudov, G, Garringer, H.J, Vidal, R, Crowther, R.A, Ghetti, B, Scheres, S.H.W, Goedert, M. | | Deposit date: | 2018-06-26 | | Release date: | 2018-09-12 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structures of filaments from Pick's disease reveal a novel tau protein fold.
Nature, 561, 2018
|
|
1GUW
 
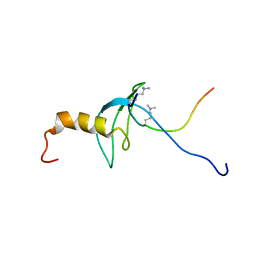 | | STRUCTURE OF THE CHROMODOMAIN FROM MOUSE HP1beta IN COMPLEX WITH THE LYSINE 9-METHYL HISTONE H3 N-TERMINAL PEPTIDE, NMR, 25 STRUCTURES | | Descriptor: | CHROMOBOX PROTEIN HOMOLOG 1, HISTONE H3.1 | | Authors: | Nielsen, P.R, Nietlispach, D, Mott, H.R, Callaghan, J.M, Bannister, A, Kouzarides, T, Murzin, A.G, Murzina, N.V, Laue, E.D. | | Deposit date: | 2002-02-01 | | Release date: | 2002-03-12 | | Last modified: | 2018-01-17 | | Method: | SOLUTION NMR | | Cite: | Structure of the Hp1 Chromodomain Bound to Histone H3 Methylated at Lysine 9
Nature, 416, 2002
|
|
7NRV
 
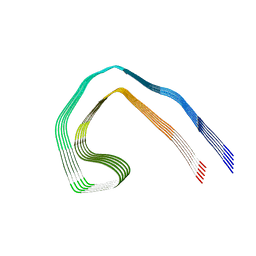 | | Paired helical filament from Alzheimer's disease with PET ligand APN-1607 | | Descriptor: | Microtubule-associated protein tau | | Authors: | Shi, Y, Murzin, A.G, Falcon, B, Epstein, A, Machin, J, Tempest, P, Newell, K.L, Vidal, R, Garringer, H.J, Sahara, N, Higuchi, M, Ghetti, B, Jang, M, Scheres, S.H.W, Goedert, M. | | Deposit date: | 2021-03-04 | | Release date: | 2021-03-24 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Cryo-EM structures of tau filaments from Alzheimer's disease with PET ligand APN-1607.
Acta Neuropathol, 141, 2021
|
|
7NRS
 
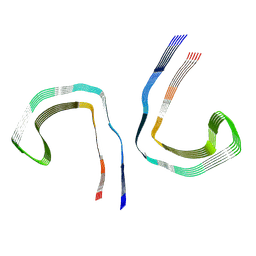 | | Conformation 1 of straight filament from primary age-related tauopathy brain | | Descriptor: | Microtubule-associated protein tau | | Authors: | Shi, Y, Murzin, A.G, Falcon, B, Epstein, A, Machin, J, Tempest, P, Newell, K.L, Vidal, R, Garringer, H.J, Sahara, N, Higuchi, M, Ghetti, B, Jang, M, Scheres, S.H.W, Goedert, M. | | Deposit date: | 2021-03-04 | | Release date: | 2021-03-24 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (2.68 Å) | | Cite: | Cryo-EM structures of tau filaments from Alzheimer's disease with PET ligand APN-1607.
Acta Neuropathol, 141, 2021
|
|
7NRQ
 
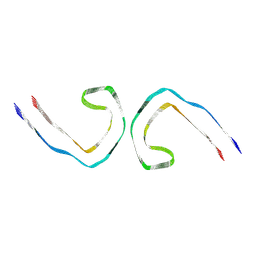 | | Paired helical filament from primary age-related tauopathy brain | | Descriptor: | Microtubule-associated protein tau | | Authors: | Shi, Y, Murzin, A.G, Falcon, B, Epstein, A, Machin, J, Tempest, P, Newell, K.L, Vidal, R, Garringer, H.J, Sahara, N, Higuchi, M, Ghetti, B, Jang, M, Scheres, S.H.W, Goedert, M. | | Deposit date: | 2021-03-04 | | Release date: | 2021-03-24 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (2.76 Å) | | Cite: | Cryo-EM structures of tau filaments from Alzheimer's disease with PET ligand APN-1607.
Acta Neuropathol, 141, 2021
|
|
7NRT
 
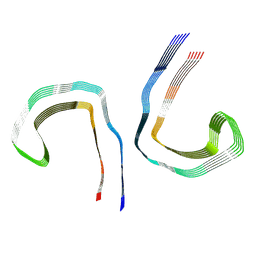 | | Conformation 2 of straight filament from primary age-related tauopathy brain | | Descriptor: | Microtubule-associated protein tau | | Authors: | Shi, Y, Murzin, A.G, Falcon, B, Epstein, A, Machin, J, Tempest, P, Newell, K.L, Vidal, R, Garringer, H.J, Sahara, N, Higuchi, M, Ghetti, B, Jang, M, Scheres, S.H.W, Goedert, M. | | Deposit date: | 2021-03-04 | | Release date: | 2021-03-24 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (2.68 Å) | | Cite: | Cryo-EM structures of tau filaments from Alzheimer's disease with PET ligand APN-1607.
Acta Neuropathol, 141, 2021
|
|
7NRX
 
 | | Straight filament from Alzheimer's disease with PET ligand APN-1607 | | Descriptor: | Microtubule-associated protein tau | | Authors: | Shi, Y, Murzin, A.G, Falcon, B, Epstein, A, Machin, J, Tempest, P, Newell, K.L, Vidal, R, Garringer, H.J, Sahara, N, Higuchi, M, Ghetti, B, Jang, M, Scheres, S.H.W, Goedert, M. | | Deposit date: | 2021-03-04 | | Release date: | 2021-03-24 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.55 Å) | | Cite: | Cryo-EM structures of tau filaments from Alzheimer's disease with PET ligand APN-1607.
Acta Neuropathol, 141, 2021
|
|
7P68
 
 | | Globular glial tauopathy type 3 tau filament | | Descriptor: | Microtubule-associated protein tau | | Authors: | Shi, Y, Zhang, W, Yang, Y, Murzin, A.G, Falcon, B, Kotecha, A, van Beers, M, Tarutani, A, Kametani, F, Garringer, H.J, Vidal, R, Hallinan, G.I, Lashley, T, Saito, Y, Murayama, S, Yoshida, M, Tanaka, H, Kakita, A, Ikeuchi, T, Robinson, A.C, Mann, D.M.A, Kovacs, G.G, Revesz, T, Ghetti, B, Hasegawa, M, Goedert, M, Scheres, S.H.W. | | Deposit date: | 2021-07-15 | | Release date: | 2021-09-15 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structure-based classification of tauopathies.
Nature, 598, 2021
|
|
7P6D
 
 | | Argyrophilic grain disease type 1 tau filament | | Descriptor: | Microtubule-associated protein tau | | Authors: | Shi, Y, Zhang, W, Yang, Y, Murzin, A.G, Falcon, B, Kotecha, A, van Beers, M, Tarutani, A, Kametani, F, Garringer, H.J, Vidal, R, Hallinan, G.I, Lashley, T, Saito, Y, Murayama, S, Yoshida, M, Tanaka, H, Kakita, A, Ikeuchi, T, Robinson, A.C, Mann, D.M.A, Kovacs, G.G, Revesz, T, Ghetti, B, Hasegawa, M, Goedert, M, Scheres, S.H.W. | | Deposit date: | 2021-07-15 | | Release date: | 2021-09-15 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structure-based classification of tauopathies.
Nature, 598, 2021
|
|
7P67
 
 | | Globular glial tauopathy type 2 tau filament | | Descriptor: | Microtubule-associated protein tau | | Authors: | Shi, Y, Zhang, W, Yang, Y, Murzin, A.G, Falcon, B, Kotecha, A, van Beers, M, Tarutani, A, Kametani, F, Garringer, H.J, Vidal, R, Hallinan, G.I, Lashley, T, Saito, Y, Murayama, S, Yoshida, M, Tanaka, H, Kakita, A, Ikeuchi, T, Robinson, A.C, Mann, D.M.A, Kovacs, G.G, Revesz, T, Ghetti, B, Hasegawa, M, Goedert, M, Scheres, S.H.W. | | Deposit date: | 2021-07-15 | | Release date: | 2021-09-15 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structure-based classification of tauopathies.
Nature, 598, 2021
|
|
7P6E
 
 | | Argyrophilic grain disease type 2 tau filament | | Descriptor: | Microtubule-associated protein tau | | Authors: | Shi, Y, Zhang, W, Yang, Y, Murzin, A.G, Falcon, B, Kotecha, A, van Beers, M, Tarutani, A, Kametani, F, Garringer, H.J, Vidal, R, Hallinan, G.I, Lashley, T, Saito, Y, Murayama, S, Yoshida, M, Tanaka, H, Kakita, A, Ikeuchi, T, Robinson, A.C, Mann, D.M.A, Kovacs, G.G, Revesz, T, Ghetti, B, Hasegawa, M, Goedert, M, Scheres, S.H.W. | | Deposit date: | 2021-07-15 | | Release date: | 2021-09-15 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structure-based classification of tauopathies.
Nature, 598, 2021
|
|
