1HP4
 
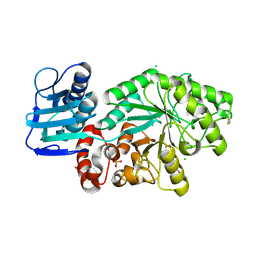 | |
1HP5
 
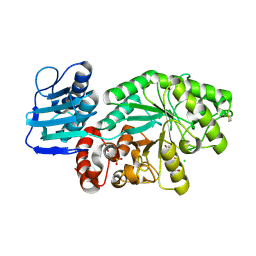 | | STREPTOMYCES PLICATUS BETA-N-ACETYLHEXOSAMINIDASE COMPLEXED WITH INTERMEDIATE ANALOUGE NAG-THIAZOLINE | | Descriptor: | 3AR,5R,6S,7R,7AR-5-HYDROXYMETHYL-2-METHYL-5,6,7,7A-TETRAHYDRO-3AH-PYRANO[3,2-D]THIAZOLE-6,7-DIOL, BETA-N-ACETYLHEXOSAMINIDASE, CHLORIDE ION, ... | | Authors: | Mark, B.L. | | Deposit date: | 2000-12-12 | | Release date: | 2001-04-04 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystallographic evidence for substrate-assisted catalysis in a bacterial beta-hexosaminidase.
J.Biol.Chem., 276, 2001
|
|
1JAK
 
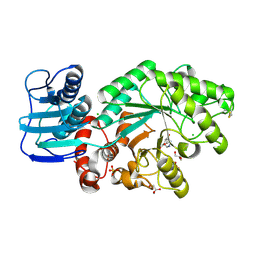 | | Streptomyces plicatus beta-N-acetylhexosaminidase in Complex with (2R,3R,4S,5R)-2-acetamido-3,4-dihydroxy-5-hydroxymethyl-piperidinium chloride (IFG) | | Descriptor: | (2R,3R,4S,5R)-2-ACETAMIDO-3,4-DIHYDROXY-5-HYDROXYMETHYL-PIPERIDINE, Beta-N-acetylhexosaminidase, CHLORIDE ION, ... | | Authors: | Mark, B.L, Vocadlo, D.J, Zhao, D, Knapp, S, Withers, S.G, James, M.N. | | Deposit date: | 2001-05-30 | | Release date: | 2001-11-21 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Biochemical and structural assessment of the 1-N-azasugar GalNAc-isofagomine as a potent family 20 beta-N-acetylhexosaminidase inhibitor.
J.Biol.Chem., 276, 2001
|
|
1NP0
 
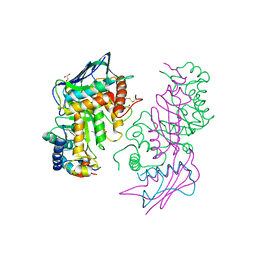 | | Human lysosomal beta-hexosaminidase isoform B in complex with intermediate analogue NAG-thiazoline | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 3AR,5R,6S,7R,7AR-5-HYDROXYMETHYL-2-METHYL-5,6,7,7A-TETRAHYDRO-3AH-PYRANO[3,2-D]THIAZOLE-6,7-DIOL, Beta-hexosaminidase subunit beta, ... | | Authors: | Mark, B.L, Mahuran, D.J, Cherney, M.M, Zhao, D, Knapp, S, James, M.N.G. | | Deposit date: | 2003-01-16 | | Release date: | 2003-04-29 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of Human beta-hexosaminidase B: Understanding the molecular basis of Sandhoff and Tay-Sachs disease
J.Mol.Biol., 327, 2003
|
|
1NOU
 
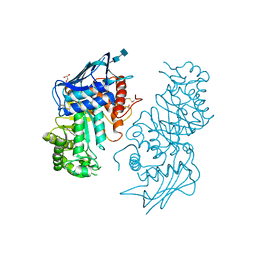 | | Native human lysosomal beta-hexosaminidase isoform B | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, SULFATE ION, ... | | Authors: | Mark, B.L, Mahuran, D.J, Cherney, M.M, Zhao, D, Knapp, S, James, M.N.G. | | Deposit date: | 2003-01-16 | | Release date: | 2003-04-08 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of Human beta-hexosaminidase B: Understanding the molecular basis of Sandhoff and Tay-Sachs disease
J.Mol.Biol., 327, 2003
|
|
1NOW
 
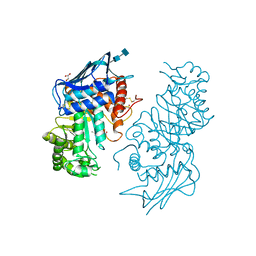 | | Human lysosomal beta-hexosaminidase isoform B in complex with (2R,3R,4S,5R)-2-Acetamido-3,4-Dihydroxy-5-Hydroxymethyl-Piperidinium Chloride (GalNAc-isofagomine) | | Descriptor: | (2R,3R,4S,5R)-2-ACETAMIDO-3,4-DIHYDROXY-5-HYDROXYMETHYL-PIPERIDINE, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, ... | | Authors: | Mark, B.L, Mahuran, D.J, Cherney, M.M, Zhao, D, Knapp, S, James, M.N.G. | | Deposit date: | 2003-01-16 | | Release date: | 2003-04-29 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of Human beta-hexosaminidase B: Understanding the molecular basis of Sandhoff and Tay-Sachs disease
J.Mol.Biol., 327, 2003
|
|
3KOT
 
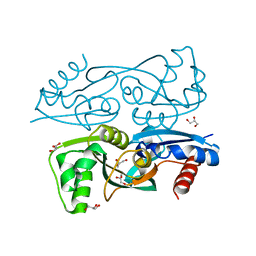 | |
6MK6
 
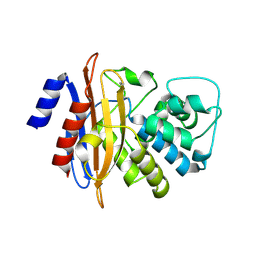 | |
6MKQ
 
 | | Carbapenemase VCC-1 bound to avibactam | | Descriptor: | (2S,5R)-1-formyl-5-[(sulfooxy)amino]piperidine-2-carboxamide, Beta-lactamase | | Authors: | Mark, B.L, Vadlamani, G. | | Deposit date: | 2018-09-26 | | Release date: | 2019-01-30 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Molecular Basis for the Potent Inhibition of the Emerging Carbapenemase VCC-1 by Avibactam.
Antimicrob. Agents Chemother., 63, 2019
|
|
3KOS
 
 | |
6DTE
 
 | | GlcNAc-inspired cyclophellitol bound to NagZ | | Descriptor: | 2,2,2-trifluoro-N-[(1R,2R,3R,4R,5R,6R)-2,3,5,6-tetrahydroxy-4-(hydroxymethyl)cyclohexyl]acetamide, Beta-hexosaminidase | | Authors: | Mark, B.L, Winogrodzki, J.L. | | Deposit date: | 2018-06-15 | | Release date: | 2019-04-24 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.929 Å) | | Cite: | A mechanism-based GlcNAc-inspired cyclophellitol inactivator of the peptidoglycan recycling enzyme NagZ reverses resistance to beta-lactams in Pseudomonas aeruginosa.
Chem. Commun. (Camb.), 54, 2018
|
|
6PEI
 
 | | Structure of sorbitol dehydrogenase from Sinorhizobium meliloti 1021 | | Descriptor: | Sorbitol dehydrogenase (L-iditol 2-dehydrogenase) | | Authors: | Bailey-Elkin, B.A, Kohlmeier, M.G, Oresnik, I.J, Mark, B.L. | | Deposit date: | 2019-06-20 | | Release date: | 2020-06-24 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Characterization of the sorbitol dehydrogenase SmoS from Sinorhizobium meliloti 1021
Acta Crystallogr.,Sect.D, 77, 2021
|
|
6PEJ
 
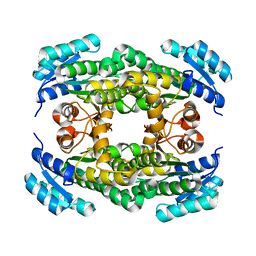 | | Structure of sorbitol dehydrogenase from Sinorhizobium meliloti 1021 bound to sorbitol | | Descriptor: | Sorbitol dehydrogenase (L-iditol 2-dehydrogenase), sorbitol | | Authors: | Bailey-Elkin, B.A, Kohlmeier, M.G, Oresnik, I.J, Mark, B.L. | | Deposit date: | 2019-06-20 | | Release date: | 2020-06-24 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Characterization of the sorbitol dehydrogenase SmoS from Sinorhizobium meliloti 1021
Acta Crystallogr.,Sect.D, 77, 2021
|
|
5FD0
 
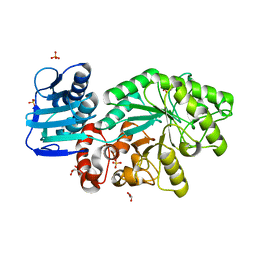 | |
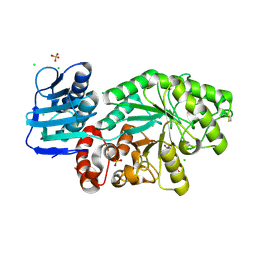
5FCZ
 
 | |
4WKM
 
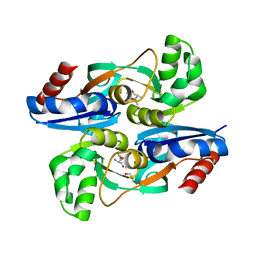 | | AmpR effector binding domain from Citrobacter freundii bound to UDP-MurNAc-pentapeptide | | Descriptor: | ALA-FGA-API-DAL-DAL, GLYCEROL, LysR family transcriptional regulator, ... | | Authors: | Vadlamani, G, Reeve, T.M, Mark, B.L. | | Deposit date: | 2014-10-02 | | Release date: | 2014-12-17 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | The beta-Lactamase Gene Regulator AmpR Is a Tetramer That Recognizes and Binds the d-Ala-d-Ala Motif of Its Repressor UDP-N-acetylmuramic Acid (MurNAc)-pentapeptide.
J.Biol.Chem., 290, 2015
|
|
8CX9
 
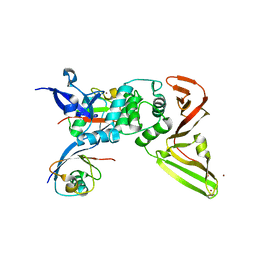 | | Structure of the SARS-COV2 PLpro (C111S) in complex with a dimeric Ubv that inhibits activity by an unusual allosteric mechanism | | Descriptor: | BROMIDE ION, CHLORIDE ION, Papain-like protease nsp3, ... | | Authors: | Singer, A.U, Slater, C.L, Patel, A, Russel, R, Mark, B.L, Sidhu, S.S. | | Deposit date: | 2022-05-20 | | Release date: | 2023-01-25 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Ubiquitin variants potently inhibit SARS-CoV-2 PLpro and viral replication via a novel site distal to the protease active site.
Plos Pathog., 18, 2022
|
|
7MIA
 
 | | Maize rayado fino virus protease | | Descriptor: | RNA replication protein | | Authors: | Patel, A, Mark, B.L. | | Deposit date: | 2021-04-16 | | Release date: | 2021-07-21 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The endopeptidase of the maize-affecting Marafivirus type member maize rayado fino virus doubles as a deubiquitinase.
J.Biol.Chem., 297, 2021
|
|
7MIC
 
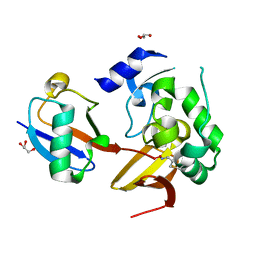 | | Maize rayado fino virus protease in complex with Ubiquitin | | Descriptor: | 3-AMINOPROPANE, GLYCEROL, RNA replication protein, ... | | Authors: | Patel, A, Mark, B.L. | | Deposit date: | 2021-04-16 | | Release date: | 2021-07-21 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | The endopeptidase of the maize-affecting Marafivirus type member maize rayado fino virus doubles as a deubiquitinase.
J.Biol.Chem., 297, 2021
|
|
8EHO
 
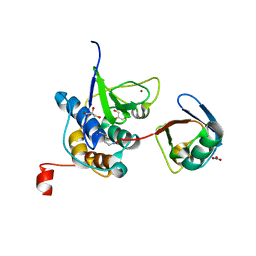 | | PRRSV-1 PLP2 domain bound to ubiquitin | | Descriptor: | 3-AMINOPROPANE, GLYCEROL, NITRATE ION, ... | | Authors: | Bailey-Elkin, B.A, Mark, B.L. | | Deposit date: | 2022-09-14 | | Release date: | 2023-12-06 | | Last modified: | 2024-01-03 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Demonstrating the importance of porcine reproductive and respiratory syndrome virus papain-like protease 2 deubiquitinating activity in viral replication by structure-guided mutagenesis.
Plos Pathog., 19, 2023
|
|
8EHN
 
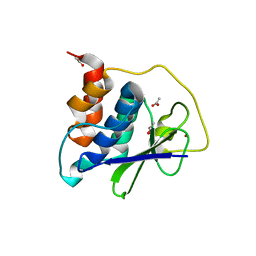 | | PRRSV-1 PLP2 domain | | Descriptor: | ACETATE ION, Papain-like protease 2, ZINC ION | | Authors: | Bailey-Elkin, B.A, Mark, B.L. | | Deposit date: | 2022-09-14 | | Release date: | 2023-12-06 | | Last modified: | 2024-01-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Demonstrating the importance of porcine reproductive and respiratory syndrome virus papain-like protease 2 deubiquitinating activity in viral replication by structure-guided mutagenesis.
Plos Pathog., 19, 2023
|
|
5DN5
 
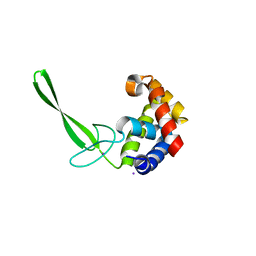 | | Structure of a C-terminally truncated glycoside hydrolase domain from Salmonella typhimurium FlgJ | | Descriptor: | CHLORIDE ION, IODIDE ION, Peptidoglycan hydrolase FlgJ, ... | | Authors: | Zaloba, P, Bailey-Elkin, B.A, Mark, B.L. | | Deposit date: | 2015-09-09 | | Release date: | 2016-02-24 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural and Biochemical Insights into the Peptidoglycan Hydrolase Domain of FlgJ from Salmonella typhimurium.
Plos One, 11, 2016
|
|
5DN4
 
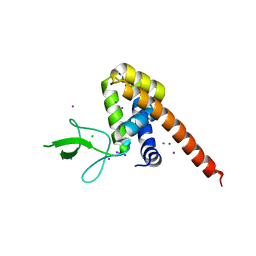 | |
4IUM
 
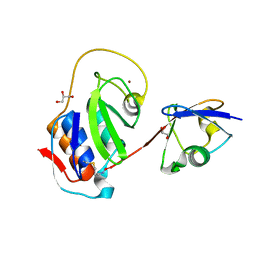 | | Equine arteritis virus papain-like protease 2 (PLP2) covalently bound to ubiquitin | | Descriptor: | GLYCEROL, Ubiquitin, ZINC ION, ... | | Authors: | Bailey-Elkin, B.A, James, T.W, Mark, B.L. | | Deposit date: | 2013-01-21 | | Release date: | 2013-02-13 | | Last modified: | 2013-03-20 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Deubiquitinase function of arterivirus papain-like protease 2 suppresses the innate immune response in infected host cells.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4GYK
 
 | |
