7HVP
 
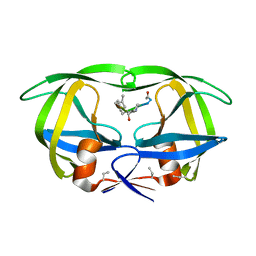 | | X-RAY CRYSTALLOGRAPHIC STRUCTURE OF A COMPLEX BETWEEN A SYNTHETIC PROTEASE OF HUMAN IMMUNODEFICIENCY VIRUS 1 AND A SUBSTRATE-BASED HYDROXYETHYLAMINE INHIBITOR | | Descriptor: | HIV-1 PROTEASE, INHIBITOR ACE-SER-LEU-ASN-PHE-PSI(CH(OH)-CH2N)-PRO-ILE VME (JG-365) | | Authors: | Swain, A.L, Miller, M.M, Green, J, Rich, D.H, Schneider, J, Kent, S.B.H, Wlodawer, A. | | Deposit date: | 1990-09-13 | | Release date: | 1993-07-15 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | X-ray crystallographic structure of a complex between a synthetic protease of human immunodeficiency virus 1 and a substrate-based hydroxyethylamine inhibitor.
Proc.Natl.Acad.Sci.USA, 87, 1990
|
|
8HVP
 
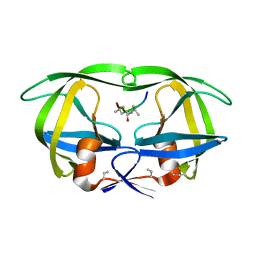 | | STRUCTURE AT 2.5-ANGSTROMS RESOLUTION OF CHEMICALLY SYNTHESIZED HUMAN IMMUNODEFICIENCY VIRUS TYPE 1 PROTEASE COMPLEXED WITH A HYDROXYETHYLENE*-BASED INHIBITOR | | Descriptor: | HIV-1 PROTEASE, INHIBITOR VAL-SER-GLN-ASN-LEU-PSI(CH(OH)-CH2)-VAL-ILE-VAL (U-85548E) | | Authors: | Jaskolski, M, Miller, M, Tomasselli, A.G, Sawyer, T.K, Staples, D.G, Heinrikson, R.L, Schneider, J, Kent, S.B.H, Wlodawer, A. | | Deposit date: | 1990-10-26 | | Release date: | 1993-10-31 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure at 2.5-A resolution of chemically synthesized human immunodeficiency virus type 1 protease complexed with a hydroxyethylene-based inhibitor.
Biochemistry, 30, 1991
|
|
4HVP
 
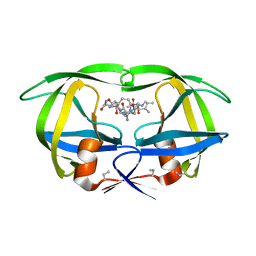 | | Structure of complex of synthetic HIV-1 protease with a substrate-based inhibitor at 2.3 Angstroms resolution | | Descriptor: | HIV-1 PROTEASE, N-{(2S)-2-[(N-acetyl-L-threonyl-L-isoleucyl)amino]hexyl}-L-norleucyl-L-glutaminyl-N~5~-[amino(iminio)methyl]-L-ornithinamide | | Authors: | Miller, M, Schneider, J, Sathyanarayana, B.K, Toth, M.V, Marshall, G.R, Clawson, L, Selk, L, Kent, S.B.H, Wlodawer, A. | | Deposit date: | 1989-08-08 | | Release date: | 1990-04-15 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of complex of synthetic HIV-1 protease with a substrate-based inhibitor at 2.3 A resolution.
Science, 246, 1989
|
|
7LL8
 
 | | D-Protein RFX-V1 Bound to the VEGFR1 Domain 2 Site on VEGF-A | | Descriptor: | Isoform L-VEGF189 of Vascular endothelial growth factor A, RFX-V1 | | Authors: | Marinec, P.S, Landgraf, K.E, Uppalapati, M, Chen, G, Xie, D, Jiang, Q, Zhao, Y, Petriello, A, Deshayes, K, Kent, S.B.H, Ault-Riche, D, Sidhu, S.S. | | Deposit date: | 2021-02-03 | | Release date: | 2021-02-17 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | A Non-immunogenic Bivalent d-Protein Potently Inhibits Retinal Vascularization and Tumor Growth.
Acs Chem.Biol., 16, 2021
|
|
7LL9
 
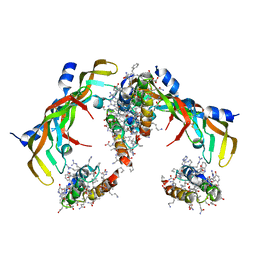 | | D-Protein RFX-V2 Bound to the VEGFR1 Domain 3 Site on VEGF-A | | Descriptor: | Isoform L-VEGF189 of Vascular endothelial growth factor A, RFX-V2 | | Authors: | Marinec, P.S, Landgraf, K.E, Uppalapati, M, Chen, G, Xie, D, Jiang, Q, Zhao, Y, Petriello, A, Deshayes, K, Kent, S.B.H, Ault-Riche, D, Sidhu, S.S. | | Deposit date: | 2021-02-03 | | Release date: | 2021-03-03 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | A Non-immunogenic Bivalent d-Protein Potently Inhibits Retinal Vascularization and Tumor Growth.
Acs Chem.Biol., 16, 2021
|
|
3DCK
 
 | | X-ray structure of D25N chemical analogue of HIV-1 protease complexed with ketomethylene isostere inhibitor | | Descriptor: | (2S)-2-{[(2R,5S)-5-{[(2S,3S)-2-{[(2S,3R)-2-(acetylamino)-3-hydroxybutanoyl]amino}-3-methylpentanoyl]amino}-2-butyl-4-oxononanoyl]amino}-N~1~-[(2S)-1-amino-5-carbamimidamido-1-oxopentan-2-yl]pentanediamide, Chemical analogue HIV-1 protease | | Authors: | Torbeev, V.Y, Mandal, K, Terechko, V.A, Kent, S.B.H. | | Deposit date: | 2008-06-03 | | Release date: | 2008-08-19 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of chemically synthesized HIV-1 protease and a ketomethylene isostere inhibitor based on the p2/NC cleavage site
Bioorg.Med.Chem.Lett., 18, 2008
|
|
3DCR
 
 | | X-ray structure of HIV-1 protease and hydrated form of ketomethylene isostere inhibitor | | Descriptor: | Chemical analogue HIV-1 protease, N~2~-[(2R,5S)-5-({(2S,3S)-2-[(N-acetyl-L-threonyl)amino]-3-methylpent-4-enoyl}amino)-2-butyl-4,4-dihydroxynonanoyl]-L-glutaminyl-L-argininamide | | Authors: | Torbeev, V.Y, Mandal, K, Terechko, V.A, Kent, S.B.H. | | Deposit date: | 2008-06-04 | | Release date: | 2008-08-19 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal structure of chemically synthesized HIV-1 protease and a ketomethylene isostere inhibitor based on the p2/NC cleavage site
Bioorg.Med.Chem.Lett., 18, 2008
|
|
2OQF
 
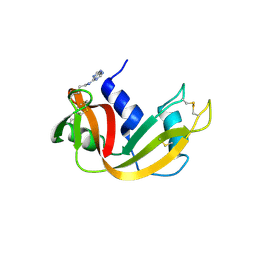 | | Structure of a synthetic, non-natural analogue of RNase A: [N71K(Ade), D83A]RNase A | | Descriptor: | Ribonuclease pancreatic | | Authors: | Boerema, D.J, Tereshko, V.A, Zhang, J.L, He, C, Kent, S.B.H. | | Deposit date: | 2007-01-31 | | Release date: | 2008-02-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Design, Synthesis, and Characterization of Non-natural RNase A Analogues with Enhanced Second-step Catalytic Activity
To be Published
|
|
3IA9
 
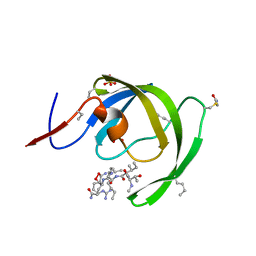 | |
3HZC
 
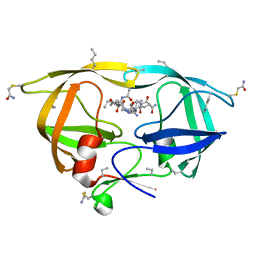 | |
3I2L
 
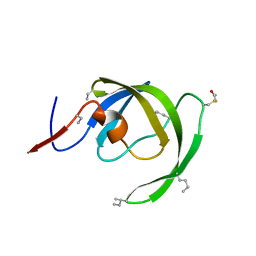 | |
3GI0
 
 | |
1HYK
 
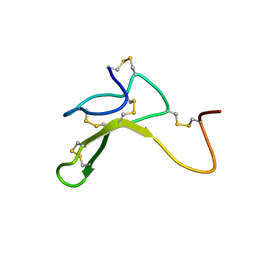 | | AGOUTI-RELATED PROTEIN (87-132) (AC-AGRP(87-132)) | | Descriptor: | AGOUTI RELATED PROTEIN | | Authors: | Bolin, K.A, Anderson, D.J, Trulson, J.A, Thompson, D.A, Wilken, J, Kent, S.B.H, Millhauser, G.L. | | Deposit date: | 2001-01-19 | | Release date: | 2001-02-07 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | NMR structure of a minimized human agouti related protein prepared by total chemical synthesis.
FEBS Lett., 451, 1999
|
|
3UE7
 
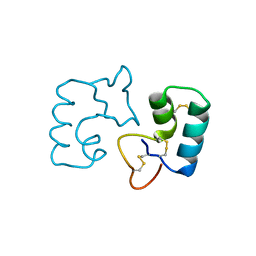 | | X-ray crystal structure of a novel topological analogue of crambin | | Descriptor: | Crambin, D-Crambin | | Authors: | Mandal, K, Pentelute, B.L, Bang, D, Gates, Z.P, Torbeev, V.Y, Kent, S.B.H. | | Deposit date: | 2011-10-28 | | Release date: | 2012-02-08 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.08 Å) | | Cite: | Design, total chemical synthesis, and x-ray structure of a protein having a novel linear-loop polypeptide chain topology.
Angew.Chem.Int.Ed.Engl., 51, 2012
|
|
6KMP
 
 | | 100K X-ray structure of HIV-1 protease triple mutant (V32I,I47V,V82I) with tetrahedral intermediate mimic KVS-1 | | Descriptor: | N~2~-[(2R,5S)-5-({(2S,3S)-2-[(N-acetyl-L-threonyl)amino]-3-methylpent-4-enoyl}amino)-2-butyl-4,4-dihydroxynonanoyl]-L-glutaminyl-L-argininamide, Protease | | Authors: | Das, A, Kovalevsky, A. | | Deposit date: | 2019-07-31 | | Release date: | 2020-07-29 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.31 Å) | | Cite: | Visualizing Tetrahedral Oxyanion Bound in HIV-1 Protease Using Neutrons: Implications for the Catalytic Mechanism and Drug Design.
Acs Omega, 5, 2020
|
|
4LFQ
 
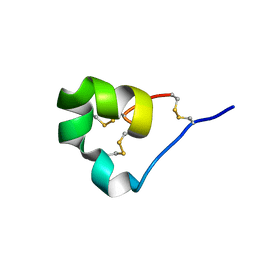 | | High resolution x-ray crystal structure of L-ShK toxin | | Descriptor: | Potassium channel toxin ShK | | Authors: | Dang, B, Kubota, T, Mandal, K, Bezanilla, F, Kent, S.B.H. | | Deposit date: | 2013-06-27 | | Release date: | 2013-08-14 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (1.059 Å) | | Cite: | Native Chemical Ligation at Asx-Cys, Glx-Cys: Chemical Synthesis and High-Resolution X-ray Structure of ShK Toxin by Racemic Protein Crystallography.
J.Am.Chem.Soc., 135, 2013
|
|
4LFS
 
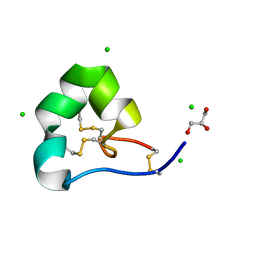 | | High resolution x-ray structure of racemic ShK toxin | | Descriptor: | CHLORIDE ION, GLYCEROL, Potassium channel toxin ShK | | Authors: | Dang, B, Kubota, T, Mandal, K, Bezanilla, F, Kent, S.B.H. | | Deposit date: | 2013-06-27 | | Release date: | 2013-08-14 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (0.97 Å) | | Cite: | Native Chemical Ligation at Asx-Cys, Glx-Cys: Chemical Synthesis and High-Resolution X-ray Structure of ShK Toxin by Racemic Protein Crystallography.
J.Am.Chem.Soc., 135, 2013
|
|
4WPY
 
 | | Racemic crystal structure of Rv1738 from Mycobacterium tuberculosis (Form-II) | | Descriptor: | GLYCEROL, protein DL-Rv1738, trifluoroacetic acid | | Authors: | Bunker, R.D, Mandal, K, Kent, S.B.H, Baker, E.N. | | Deposit date: | 2014-10-21 | | Release date: | 2015-03-18 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | A functional role of Rv1738 in Mycobacterium tuberculosis persistence suggested by racemic protein crystallography.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
4WSP
 
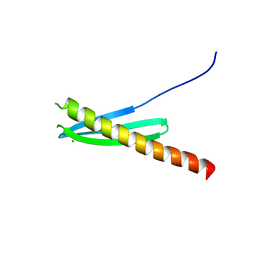 | | Racemic crystal structure of Rv1738 from Mycobacterium tuberculosis (Form-I) | | Descriptor: | CHLORIDE ION, protein DL-Rv1738 | | Authors: | Bunker, R.D, Mandal, K, Kent, S.B.H, Baker, E.N. | | Deposit date: | 2014-10-28 | | Release date: | 2015-03-18 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | A functional role of Rv1738 in Mycobacterium tuberculosis persistence suggested by racemic protein crystallography.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
1B3A
 
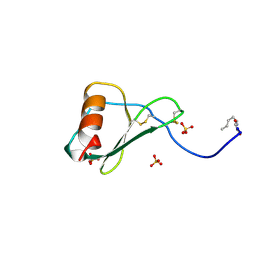 | | TOTAL CHEMICAL SYNTHESIS AND HIGH-RESOLUTION CRYSTAL STRUCTURE OF THE POTENT ANTI-HIV PROTEIN AOP-RANTES | | Descriptor: | PENTYLOXYAMINO-ACETALDEHYDE, PROTEIN (RANTES), SULFATE ION | | Authors: | Wilken, J, Hoover, D, Thompson, D.A, Barlow, P.N, Mcsparron, H, Picard, L, Wlodawer, A, Lubkowski, J, Kent, S.B.H. | | Deposit date: | 1998-12-07 | | Release date: | 1999-04-23 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Total chemical synthesis and high-resolution crystal structure of the potent anti-HIV protein AOP-RANTES.
Chem.Biol., 6, 1999
|
|
4PHV
 
 | | X-RAY CRYSTAL STRUCTURE OF THE HIV PROTEASE COMPLEX WITH L-700,417, AN INHIBITOR WITH PSEUDO C2 SYMMETRY | | Descriptor: | HIV-1 PROTEASE, N,N-BIS(2-HYDROXY-1-INDANYL)-2,6- DIPHENYLMETHYL-4-HYDROXY-1,7-HEPTANDIAMIDE | | Authors: | Bone, R. | | Deposit date: | 1991-10-04 | | Release date: | 1993-10-31 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | X-Ray Crystal Structure of the HIV Protease Complex with L-700,417, an Inhibitor with Pseudo C2 Symmetry
J.Am.Chem.Soc., 113, 1991
|
|
4IUZ
 
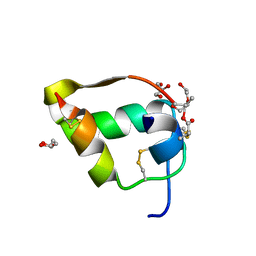 | | High resolution crystal structure of racemic ester insulin | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, Insulin A chain, ... | | Authors: | Avital-Shmilovici, M, Mandal, K, Gates, Z.P, Phillips, N, Weiss, M.A, Kent, S.B.H. | | Deposit date: | 2013-01-22 | | Release date: | 2013-02-13 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Fully Convergent Chemical Synthesis of Ester Insulin: Determination of the High Resolution X-ray Structure by Racemic Protein Crystallography.
J.Am.Chem.Soc., 135, 2013
|
|
3BOG
 
 | | Snow Flea Antifreeze Protein Quasi-racemate | | Descriptor: | 6.5 kDa glycine-rich antifreeze protein, UNKNOWN LIGAND | | Authors: | Pentelute, B.L, Kent, S.B.H, Gates, Z.P, Tereshko, V, Kossiakoff, A.A, Kurutz, J, Dashnau, J, Vaderkooi, J.M. | | Deposit date: | 2007-12-17 | | Release date: | 2008-09-23 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | X-ray structure of snow flea antifreeze protein determined by racemic crystallization of synthetic protein enantiomers
J.Am.Chem.Soc., 130, 2008
|
|
5UDP
 
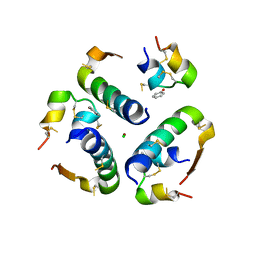 | | High resolution x-ray crystal structure of synthetic insulin lispro | | Descriptor: | CHLORIDE ION, Insulin Lispro A chain, Insulin Lispro B chain, ... | | Authors: | Mandal, K, Dhayalan, B, Kent, S.B.H. | | Deposit date: | 2016-12-28 | | Release date: | 2017-01-11 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.348 Å) | | Cite: | Scope and Limitations of Fmoc Chemistry SPPS-Based Approaches to the Total Synthesis of Insulin Lispro via Ester Insulin.
Chemistry, 23, 2017
|
|
4OIJ
 
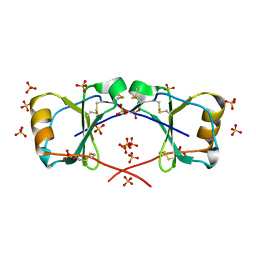 | | X-ray crystal structure of racemic non-glycosylated chemokine Ser-CCL1 | | Descriptor: | C-C motif chemokine 1, D-Ser-CCL1, SULFATE ION | | Authors: | Okamoto, R, Mandal, K, Sawaya, M.R, Kajihara, Y, Yeates, T.O, Kent, S.B.H. | | Deposit date: | 2014-01-19 | | Release date: | 2014-05-07 | | Last modified: | 2014-05-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | (Quasi-)Racemic X-ray Structures of Glycosylated and Non-Glycosylated Forms of the Chemokine Ser-CCL1 Prepared by Total Chemical Synthesis.
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
