7UFG
 
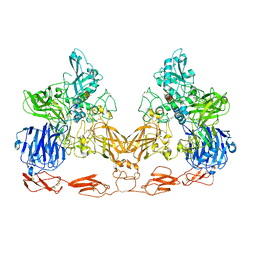 | | Cryo-EM structure of PAPP-A in complex with IGFBP5 | | Descriptor: | Insulin-like growth factor-binding protein 5, Pappalysin-1, ZINC ION | | Authors: | Judge, R.A, Jain, R, Hao, Q, Ouch, C, Sridar, J, Smith, C.L, Wang, J.C.K, Eaton, D. | | Deposit date: | 2022-03-22 | | Release date: | 2022-09-28 | | Method: | ELECTRON MICROSCOPY (3.28 Å) | | Cite: | Structure of the PAPP-ABP5 complex reveals mechanism of substrate recognition
Nat Commun, 13, 2022
|
|
8D8O
 
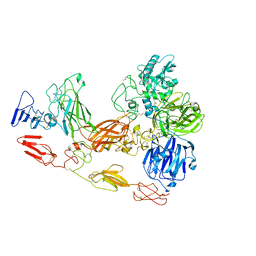 | | Cryo-EM structure of substrate unbound PAPP-A | | Descriptor: | Pappalysin-1, ZINC ION | | Authors: | Judge, R.A, Jain, R, Hao, Q, Ouch, C, Sridar, J, Smith, C.L, Wang, J.C.K, Eaton, D. | | Deposit date: | 2022-06-08 | | Release date: | 2022-09-28 | | Method: | ELECTRON MICROSCOPY (3.35 Å) | | Cite: | Structure of the PAPP-ABP5 complex reveals mechanism of substrate recognition
Nat Commun, 13, 2022
|
|
5TEG
 
 | |
5VHB
 
 | |
5VIB
 
 | |
5VI9
 
 | |
8VKZ
 
 | | Crystal structure of Glucocorticoid Receptor in complex with an inhibitor | | Descriptor: | (4aR,4bS,5R,6aS,6bS,8R,9aR,10aR,10bR)-8-{4-[(3-aminophenyl)methyl]phenyl}-5-hydroxy-6b-(hydroxyacetyl)-4a,6a-dimethyl-4a,4b,5,6,6a,6b,9a,10,10a,10b,11,12-dodecahydro-2H,8H-naphtho[2',1':4,5]indeno[1,2-d][1,3]dioxol-2-one, Glucocorticoid receptor, Nuclear receptor coactivator 2 | | Authors: | Judge, R.A, Hobson, A.D. | | Deposit date: | 2024-01-10 | | Release date: | 2024-01-24 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.133 Å) | | Cite: | Minimising the payload solvent exposed hydrophobic surface area optimises the antibody-drug conjugate properties
Rsc Med Chem, 15, 2024
|
|
8SL1
 
 | | Cryo-EM structure of PAPP-A2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Judge, R.A, Stoll, V.S, Eaton, D, Hao, Q, Bratkowski, M.A. | | Deposit date: | 2023-04-20 | | Release date: | 2023-11-08 | | Method: | ELECTRON MICROSCOPY (3.13 Å) | | Cite: | Cryo-EM structure of human PAPP-A2 and mechanism of substrate recognition
Commun Chem, 6, 2023
|
|
7LHB
 
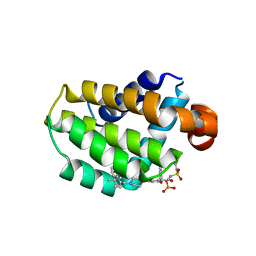 | | Crystal structure of Bcl-2 in complex with prodrug ABBV-167 | | Descriptor: | Apoptosis regulator Bcl-2, Phosphoric acid mono-[5-(5-{4-[2-(4-chloro-phenyl)-4,4-dimethyl-cyclohex-1-enylmethyl]-piperazin-1-yl}-2-{3-nitro-4-[(tetrahydro-pyran-4-ylmethyl)-amino]-benzenesulfonylaminocarbonyl}-phenoxy)-pyrrolo[2,3-b]pyridin-7-ylmethyl] ester | | Authors: | Judge, R.A, Salem, A.H. | | Deposit date: | 2021-01-21 | | Release date: | 2021-04-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.068 Å) | | Cite: | Expanding the Repertoire for "Large Small Molecules": Prodrug ABBV-167 Efficiently Converts to Venetoclax with Reduced Food Effect in Healthy Volunteers.
Mol.Cancer Ther., 20, 2021
|
|
8EL0
 
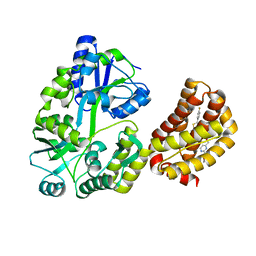 | | Structure of MBP-Mcl-1 in complex with a macrocyclic compound | | Descriptor: | (7R,20P)-18-chloro-1-(4-fluorophenyl)-10-{[(2M)-2-(2-methoxyphenyl)pyrimidin-4-yl]methoxy}-19-methyl-15-[2-(4-methylpiperazin-1-yl)ethyl]-7,8,15,16-tetrahydro-14H-17,20-etheno-9,13-(metheno)-6-oxa-2-thia-3,5,15-triazacyclooctadeca[1,2,3-cd]indene-7-carboxylic acid, Maltose/maltodextrin-binding periplasmic protein,Induced myeloid leukemia cell differentiation protein Mcl-1, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Judge, R.A, Judd, A.S, Souers, A.J. | | Deposit date: | 2022-09-22 | | Release date: | 2023-10-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.917 Å) | | Cite: | Selective MCL-1 inhibitor ABBV-467 is efficacious in tumor models but is associated with cardiac troponin increases in patients.
Commun Med (Lond), 3, 2023
|
|
8EL1
 
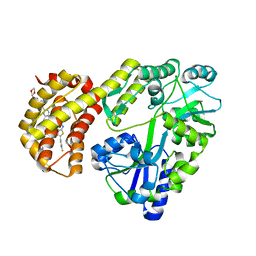 | | Structure of MBP-Mcl-1 in complex with ABBV-467 | | Descriptor: | (7R,16R)-19,23-dichloro-10-{[2-(4-{[(2R)-1,4-dioxan-2-yl]methoxy}phenyl)pyrimidin-4-yl]methoxy}-1-(4-fluorophenyl)-20,22-dimethyl-16-[(4-methylpiperazin-1-yl)methyl]-7,8,15,16-tetrahydro-18,21-etheno-13,9-(metheno)-6,14,17-trioxa-2-thia-3,5-diazacyclononadeca[1,2,3-cd]indene-7-carboxylic acid, Maltose/maltodextrin-binding periplasmic protein,Induced myeloid leukemia cell differentiation protein Mcl-1, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Judge, R.A, Judd, A.S, Souers, A.J. | | Deposit date: | 2022-09-22 | | Release date: | 2023-10-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.406 Å) | | Cite: | Selective MCL-1 inhibitor ABBV-467 is efficacious in tumor models but is associated with cardiac troponin increases in patients.
Commun Med (Lond), 3, 2023
|
|
8EKX
 
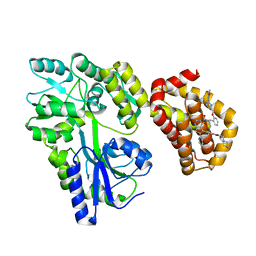 | | Structure of MBP-Mcl-1 in complex with MIK665 | | Descriptor: | (2~{R})-2-[5-[3-chloranyl-2-methyl-4-[2-(4-methylpiperazin-1-yl)ethoxy]phenyl]-6-(4-fluorophenyl)thieno[2,3-d]pyrimidin-4-yl]oxy-3-[2-[[2-(2-methoxyphenyl)pyrimidin-4-yl]methoxy]phenyl]propanoic acid, Maltose/maltodextrin-binding periplasmic protein,Induced myeloid leukemia cell differentiation protein Mcl-1, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Judge, R.A, Judd, A.S, Souers, A.J. | | Deposit date: | 2022-09-22 | | Release date: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Selective MCL-1 inhibitor ABBV-467 is efficacious in tumor models but is associated with cardiac troponin increases in patients.
Commun Med (Lond), 3, 2023
|
|
6VWC
 
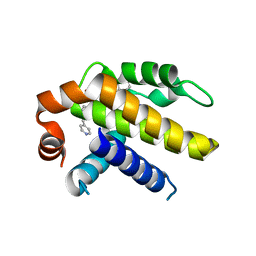 | | Crystal structure of Bcl-xL in complex with tetrahydroisoquinoline-pyridine based inhibitors | | Descriptor: | 6-{8-[(1,3-benzothiazol-2-yl)carbamoyl]-3,4-dihydroisoquinolin-2(1H)-yl}-3-{1-[(pyridin-4-yl)methyl]-1H-pyrazol-4-yl}pyridine-2-carboxylic acid, Bcl-2-like protein 1 | | Authors: | Judge, R.A, Judd, A.S. | | Deposit date: | 2020-02-19 | | Release date: | 2020-10-21 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.604 Å) | | Cite: | Discovery of A-1331852, a First-in-Class, Potent, and Orally-Bioavailable BCL-X L Inhibitor.
Acs Med.Chem.Lett., 11, 2020
|
|
6B4U
 
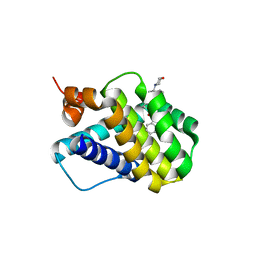 | | Crystal structure of MCL-1 in complex with a BIM competitive inhibitor | | Descriptor: | 7-(2-methylphenyl)-1-[2-(morpholin-4-yl)ethyl]-3-{3-[(naphthalen-1-yl)oxy]propyl}-1H-indole-2-carboxylic acid, Induced myeloid leukemia cell differentiation protein Mcl-1 | | Authors: | Judge, R.A, Souers, A.J. | | Deposit date: | 2017-09-27 | | Release date: | 2017-10-04 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure-guided design of a series of MCL-1 inhibitors with high affinity and selectivity.
J. Med. Chem., 58, 2015
|
|
6B4L
 
 | |
7LH7
 
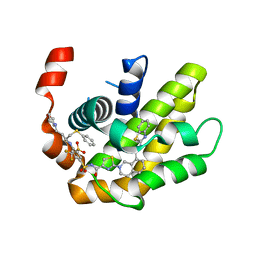 | | Crystal structure of BCL-XL in complex with a benzothiazole-based inhibitor | | Descriptor: | Bcl-2-like protein 1, N-(1,3-benzothiazol-2-yl)-2-(4-{[(4-{[(2R)-4-(morpholin-4-yl)-1-(phenylsulfanyl)butan-2-yl]amino}-3-[(trifluoromethyl)sulfonyl]phenyl)sulfonyl]carbamoyl}-1,3-thiazol-2-yl)-1,2,3,4-tetrahydroisoquinoline-8-carboxamide | | Authors: | Judge, R.A, Tao, Z. | | Deposit date: | 2021-01-21 | | Release date: | 2021-06-23 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.409 Å) | | Cite: | Structure-Based Design of A-1293102, a Potent and Selective BCL-XL Inhibitor
ACS Medicinal Chemistry Letters, 12, 2021
|
|
6E99
 
 | |
6E9W
 
 | | Crystal structure of Rock1 with a pyridinylbenzamide based inhibitor | | Descriptor: | N-[(2,3-dihydro-1,4-benzodioxin-5-yl)methyl]-4-(pyridin-4-yl)benzamide, Rho-associated protein kinase 1, SULFATE ION | | Authors: | Judge, R.A, Hobson, A.D. | | Deposit date: | 2018-08-01 | | Release date: | 2018-11-14 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.96 Å) | | Cite: | Identification of Selective Dual ROCK1 and ROCK2 Inhibitors Using Structure Based Drug Design.
J. Med. Chem., 61, 2018
|
|
6E9L
 
 | |
6ED6
 
 | |
5VAR
 
 | |
5VKC
 
 | | Crystal structure of MCL-1 in complex with a BIM competitive inhibitor | | Descriptor: | 7-(3-{[4-(4-acetylpiperazin-1-yl)phenoxy]methyl}-1,5-dimethyl-1H-pyrazol-4-yl)-3-{3-[(naphthalen-1-yl)oxy]propyl}-1-[(pyridin-3-yl)methyl]-1H-indole-2-carboxylic acid, Induced myeloid leukemia cell differentiation protein Mcl-1, ZINC ION | | Authors: | Judge, R.A, Souers, A.J. | | Deposit date: | 2017-04-21 | | Release date: | 2017-05-03 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Structure-guided design of a series of MCL-1 inhibitors with high affinity and selectivity.
J. Med. Chem., 58, 2015
|
|
5DYO
 
 | | Fab43.1 complex with flourescein | | Descriptor: | 2-(6-HYDROXY-3-OXO-3H-XANTHEN-9-YL)-BENZOIC ACID, Fab 43.1 Heavy Chain, Fab 43.1 Light Chain, ... | | Authors: | Longenecker, K.L, Judge, R.A. | | Deposit date: | 2015-09-25 | | Release date: | 2016-01-27 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Three-dimensional structure, binding, and spectroscopic characteristics of the monoclonal antibody 43.1 directed to the carboxyphenyl moiety of fluorescein.
Biopolymers, 105, 2016
|
|
5EUE
 
 | | S1P Lyase Bacterial Surrogate bound to N-(2-((4-methoxy-2,5-dimethylbenzyl)amino)-1-phenylethyl)-5-methylisoxazole-3-carboxamide | | Descriptor: | PHOSPHATE ION, Putative sphingosine-1-phosphate lyase, ~{N}-[(1~{S})-2-[(4-methoxy-2,5-dimethyl-phenyl)methylamino]-1-phenyl-ethyl]-5-methyl-1,2-oxazole-3-carboxamide | | Authors: | Argiriadi, M.A, Banach, D, Radziejewska, E, Marchie, S, DiMauro, J, Dinges, J, Dominguez, E, Hutchins, C, Judge, R.A, Queeney, K, Wallace, G, Harris, C.M. | | Deposit date: | 2015-11-18 | | Release date: | 2016-03-16 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.83 Å) | | Cite: | Creation of a S1P Lyase bacterial surrogate for structure-based drug design.
Bioorg.Med.Chem.Lett., 26, 2016
|
|
5EUD
 
 | | S1P Lyase Bacterial Surrogate bound to N-(1-(4-(3-hydroxyprop-1-yn-1-yl)phenyl)-2-((4-methoxy-2,5-dimethylbenzyl)amino)ethyl)-5-methylisoxazole-3-carboxamide | | Descriptor: | PHOSPHATE ION, Putative sphingosine-1-phosphate lyase, ~{N}-[(1~{S})-2-[(4-methoxy-2,5-dimethyl-phenyl)methylamino]-1-[4-(3-oxidanylprop-1-ynyl)phenyl]ethyl]-5-methyl-1,2-oxazole-3-carboxamide | | Authors: | Argiriadi, M.A, Banach, D, Radziejewska, E, Marchie, S, DiMauro, J, Dinges, J, Dominguez, E, Hutchins, C, Judge, R.A, Queeney, K, Wallace, G, Harris, C.M. | | Deposit date: | 2015-11-18 | | Release date: | 2016-03-16 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Creation of a S1P Lyase bacterial surrogate for structure-based drug design.
Bioorg.Med.Chem.Lett., 26, 2016
|
|
