5A0E
 
 | | Crystal structure of cyclophilin D in complex with CsA analogue, JW47. | | Descriptor: | JW47, PEPTIDYL-PROLYL CIS-TRANS ISOMERASE F, MITOCHONDRIAL | | Authors: | Warne, J, Pryce, G, Hill, J, Shi, X, Lenneras, F, Puentes, F, Kip, M, Hilditch, L, Walker, P, Simone, M, Chan, A.W.E, Towers, G, Coker, A.R, Duchen, M, Szabadkai, G, Baker, D, Selwood, D.L. | | Deposit date: | 2015-04-19 | | Release date: | 2015-12-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Selective Inhibition of the Mitochondrial Permeability Transition Pore Protects Against Neuro-Degeneration in Experimental Multiple Sclerosis.
J.Biol.Chem., 291, 2016
|
|
5BOD
 
 | | Crystal structure of Streptococcus pneumonia ParE inhibitor | | Descriptor: | (2R)-N-[3-(3,5-dimethylphenyl)-1H-pyrazol-5-yl]-1,4-dioxane-2-carboxamide, DNA topoisomerase 4 subunit B | | Authors: | Tan, Y.W, Chen, G, Hung, A.W, Hill, J. | | Deposit date: | 2015-05-27 | | Release date: | 2015-06-17 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Application of Fragment-based Drug Discovery against DNA GyraseB
to be published
|
|
5BOC
 
 | | Crystal structure of topoisomerase ParE inhibitor | | Descriptor: | 3-methyl-4-({3-[3-methyl-5-(trifluoromethyl)phenyl]-1H-pyrazol-5-yl}carbamoyl)benzoic acid, DNA topoisomerase 4 subunit B | | Authors: | Tan, Y.W, Chen, G.Y, Hung, A.W, Hill, J. | | Deposit date: | 2015-05-27 | | Release date: | 2015-06-17 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Application of Fragment-based Drug Discovery against DNA GyraseB
To be published
|
|
5YIG
 
 | | Crystal structure of Streptococcus pneumonia ParE with inhibitor | | Descriptor: | 1-ethyl-3-[5-[2-[(1S,5R)-3-methyl-3,8-diazabicyclo[3.2.1]octan-8-yl]-5-(2-oxidanylidene-3H-1,3,4-oxadiazol-5-yl)pyridin-3-yl]-4-[4-(trifluoromethyl)-1,3-thiazol-2-yl]pyridin-2-yl]urea, DNA topoisomerase 4 subunit B | | Authors: | Cherian, J, Tan, Y, Hill, J. | | Deposit date: | 2017-10-04 | | Release date: | 2018-09-05 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Discovery of dual GyrB/ParE inhibitors active against Gram-negative bacteria.
Eur J Med Chem, 157, 2018
|
|
6AFK
 
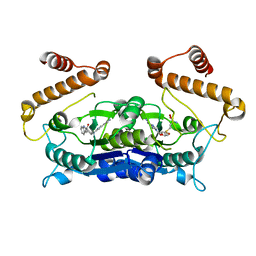 | | Crystal structure of TrmD from Pseudomonas aeruginosa in complex with active-site inhibitor | | Descriptor: | N-{(3S)-1-[3-(pyridin-4-yl)-1H-pyrazol-5-yl]piperidin-3-yl}-1H-indole-2-carboxamide, S-ADENOSYLMETHIONINE, tRNA (guanine-N(1)-)-methyltransferase | | Authors: | Zhong, W, Koay, A, Wong, Y.W, Sahili, A.E, Nah, Q, Kang, C, Poulsen, A, Chionh, Y.K, McBee, M, Matter, A, Hill, J, Lescar, J, Dedon, P.C. | | Deposit date: | 2018-08-08 | | Release date: | 2019-08-14 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Targeting the Bacterial Epitranscriptome for Antibiotic Development: Discovery of Novel tRNA-(N1G37) Methyltransferase (TrmD) Inhibitors.
Acs Infect Dis., 5, 2019
|
|
6ILZ
 
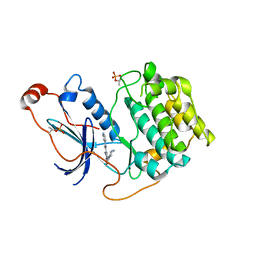 | | Crystal structure of PKCiota in complex with inhibitor | | Descriptor: | 2-amino-5-[3-(piperazin-1-yl)phenyl]-N-(pyridin-4-yl)pyridine-3-carboxamide, Protein kinase C iota type | | Authors: | Baburajendran, N, Hill, J. | | Deposit date: | 2018-10-21 | | Release date: | 2019-06-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.261 Å) | | Cite: | Fragment-based Discovery of a Small-Molecule Protein Kinase C-iota Inhibitor Binding Post-kinase Domain Residues.
Acs Med.Chem.Lett., 10, 2019
|
|
6JLR
 
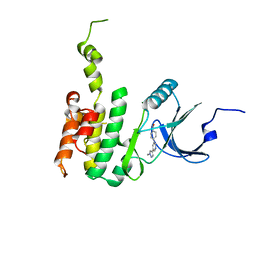 | |
7DXL
 
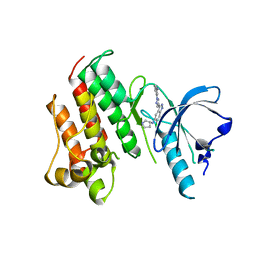 | |
7ZXK
 
 | | Human IL-27 in complex with neutralizing SRF388 FAb fragment | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Interleukin-27 subunit alpha, Interleukin-27 subunit beta, ... | | Authors: | Bloch, Y, Skladanowska, K, Strand, J, Welin, M, Logan, D, Hill, J, Savvides, S.N. | | Deposit date: | 2022-05-21 | | Release date: | 2022-11-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis of activation and antagonism of receptor signaling mediated by interleukin-27.
Cell Rep, 41, 2022
|
|
7SKR
 
 | | BtSCoV-Rf1.2004 Papain-Like protease bound to the non-covalent inhibitor 37 | | Descriptor: | 3C-like proteinase, DIMETHYL SULFOXIDE, N-[(2-methoxypyridin-4-yl)methyl]-2-[(1R)-1-(naphthalen-1-yl)ethyl]-2-azaspiro[3.3]heptane-6-carboxamide, ... | | Authors: | Durie, I, Shepard, J, Freitas, B, O'Boyle, B, Enos, S, Pegan, S.D. | | Deposit date: | 2021-10-21 | | Release date: | 2022-03-02 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | Exploring Noncovalent Protease Inhibitors for the Treatment of Severe Acute Respiratory Syndrome and Severe Acute Respiratory Syndrome-Like Coronaviruses.
Acs Infect Dis., 8, 2022
|
|
7SKQ
 
 | | BtSCoV-Rf1.2004 Papain-Like protease bound to the non-covalent inhibitor GRL-0617 | | Descriptor: | 3C-like proteinase, 5-amino-2-methyl-N-[(1R)-1-naphthalen-1-ylethyl]benzamide, ZINC ION | | Authors: | Freitas, B, Durie, I, Shepard, J, O'Boyle, B, Enos, S, Pegan, S.D. | | Deposit date: | 2021-10-21 | | Release date: | 2022-03-02 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3.16 Å) | | Cite: | Exploring Noncovalent Protease Inhibitors for the Treatment of Severe Acute Respiratory Syndrome and Severe Acute Respiratory Syndrome-Like Coronaviruses.
Acs Infect Dis., 8, 2022
|
|
4EJN
 
 | | Crystal structure of autoinhibited form of AKT1 in complex with N-(4-(5-(3-acetamidophenyl)-2-(2-aminopyridin-3-yl)-3H-imidazo[4,5-b]pyridin-3-yl)benzyl)-3-fluorobenzamide | | Descriptor: | 1,2-ETHANEDIOL, 2-BUTANOL, N-(4-{5-[3-(acetylamino)phenyl]-2-(2-aminopyridin-3-yl)-3H-imidazo[4,5-b]pyridin-3-yl}benzyl)-3-fluorobenzamide, ... | | Authors: | Eathiraj, S. | | Deposit date: | 2012-04-06 | | Release date: | 2012-05-23 | | Last modified: | 2013-01-02 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Discovery and optimization of a series of 3-(3-phenyl-3H-imidazo[4,5-b]pyridin-2-yl)pyridin-2-amines: orally bioavailable, selective, and potent ATP-independent Akt inhibitors.
J.Med.Chem., 55, 2012
|
|
3W51
 
 | | Tankyrase in complex with 2-hydroxy-4-methylquinoline | | Descriptor: | 4-methylquinolin-2-ol, SULFATE ION, Tankyrase-2, ... | | Authors: | Jansson, A.E, Larsson, E.A, Nordlund, P.L. | | Deposit date: | 2013-01-18 | | Release date: | 2013-07-10 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Fragment-based ligand design of novel potent inhibitors of tankyrases.
J.Med.Chem., 56, 2013
|
|
2KYJ
 
 | |
5MU1
 
 | | UDP-Glucose Glycoprotein Glucosyltransferase from Chaetomium thermophilum soaked with K2PtI6 | | Descriptor: | CALCIUM ION, IODIDE ION, PLATINUM (II) ION, ... | | Authors: | Roversi, P, Caputo, A.T, Hill, J, Alonzi, D.S, Zitzmann, N. | | Deposit date: | 2017-01-11 | | Release date: | 2017-07-26 | | Last modified: | 2023-03-08 | | Method: | X-RAY DIFFRACTION (3.48 Å) | | Cite: | Interdomain conformational flexibility underpins the activity of UGGT, the eukaryotic glycoprotein secretion checkpoint.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5MZO
 
 | | UDP-Glucose Glycoprotein Glucosyltransferase from Chaetomium thermophilum (open conformation) | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Roversi, P, Caputo, A.T, Hill, J, Alonzi, D.S, Zitzmann, N. | | Deposit date: | 2017-02-01 | | Release date: | 2017-07-26 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.48 Å) | | Cite: | Interdomain conformational flexibility underpins the activity of UGGT, the eukaryotic glycoprotein secretion checkpoint.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5N2J
 
 | | UDP-Glucose Glycoprotein Glucosyltransferase from Chaetomium thermophilum (closed form) | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Roversi, P, Caputo, A.T, Hill, J, Alonzi, D.S, Zitzmann, N. | | Deposit date: | 2017-02-07 | | Release date: | 2017-07-26 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (4.4 Å) | | Cite: | Interdomain conformational flexibility underpins the activity of UGGT, the eukaryotic glycoprotein secretion checkpoint.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5XWR
 
 | | Crystal Structure of RBBP4-peptide complex | | Descriptor: | Histone-binding protein RBBP4, MET-SER-ARG-ARG-LYS-GLN-ALA-LYS-PRO-GLN-HIS-ILE | | Authors: | Jobichen, C, Lui, B.H, Daniel, G.T, Sivaraman, J. | | Deposit date: | 2017-06-30 | | Release date: | 2018-07-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Targeting cancer addiction for SALL4 by shifting its transcriptome with a pharmacologic peptide.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
5ZMA
 
 | | Structural basis for an allosteric Eya2 phosphatase inhibitor | | Descriptor: | 3-fluoro-N'-[(E)-{5-[(pyrimidin-2-yl)sulfanyl]furan-2-yl}methylidene]benzohydrazide, Eyes absent homolog 2 | | Authors: | Anantharajan, J, Jansson, A.E, Kang, C. | | Deposit date: | 2018-04-02 | | Release date: | 2019-06-05 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.175 Å) | | Cite: | Structural and Functional Analyses of an Allosteric EYA2 Phosphatase Inhibitor That Has On-Target Effects in Human Lung Cancer Cells.
Mol.Cancer Ther., 18, 2019
|
|
6Y5M
 
 | | Crystal structure of mouse Autotaxin in complex with compound 1a | | Descriptor: | (~{E})-3-[4-chloranyl-2-[(5-methyl-1,2,3,4-tetrazol-2-yl)methyl]phenyl]-1-[(2~{R})-4-[(4-fluorophenyl)methyl]-2-methyl-piperazin-1-yl]prop-2-en-1-one, 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ... | | Authors: | Faller, M, Zink, F. | | Deposit date: | 2020-02-25 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.011 Å) | | Cite: | Development of autotaxin inhibitors: A series of tetrazole cinnamides.
Bioorg.Med.Chem.Lett., 31, 2021
|
|
5YJO
 
 | |
4J21
 
 | | Tankyrase 2 in complex with 7-(4-amino-2-chlorophenyl)-4-methylquinolin-2(1H)-one | | Descriptor: | 7-(4-amino-2-chlorophenyl)-4-methylquinolin-2(1H)-one, SULFATE ION, Tankyrase-2, ... | | Authors: | Jansson, A.E, Larsson, E.A, Nordlund, P.L. | | Deposit date: | 2013-02-04 | | Release date: | 2013-06-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Fragment-based ligand design of novel potent inhibitors of tankyrases.
J.Med.Chem., 56, 2013
|
|
4J3L
 
 | | Tankyrase 2 in complex with 3-chloro-N-(2-methoxyethyl)-4-(4-methyl-2-oxo-1,2-dihydroquinolin-7-yl)benzamide | | Descriptor: | 3-chloro-N-(2-methoxyethyl)-4-(4-methyl-2-oxo-1,2-dihydroquinolin-7-yl)benzamide, SULFATE ION, Tankyrase-2, ... | | Authors: | Jansson, A.E, Larsson, E.A, Nordlund, P.L. | | Deposit date: | 2013-02-05 | | Release date: | 2013-06-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Fragment-based ligand design of novel potent inhibitors of tankyrases.
J.Med.Chem., 56, 2013
|
|
4J22
 
 | | Tankyrase 2 in complex with 3-chloro-4-(4-methyl-2-oxo-1,2-dihydroquinolin-7-yl)-N-[2-(morpholin-4-yl)ethyl]benzamide | | Descriptor: | 3-chloro-4-(4-methyl-2-oxo-1,2-dihydroquinolin-7-yl)-N-[2-(morpholin-4-yl)ethyl]benzamide, SULFATE ION, Tankyrase-2, ... | | Authors: | Jansson, A.E, Larsson, E.A, Nordlund, P.L. | | Deposit date: | 2013-02-04 | | Release date: | 2013-06-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Fragment-based ligand design of novel potent inhibitors of tankyrases.
J.Med.Chem., 56, 2013
|
|
4IUE
 
 | | Tankyrase in complex with 7-(2-fluorophenyl)-4-methyl-1,2-dihydroquinolin-2-one | | Descriptor: | 7-(2-fluorophenyl)-4-methylquinolin-2(1H)-one, SULFATE ION, Tankyrase-2, ... | | Authors: | Jansson, A.E, Larsson, E.A, Nordlund, P.L. | | Deposit date: | 2013-01-21 | | Release date: | 2013-06-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Fragment-based ligand design of novel potent inhibitors of tankyrases.
J.Med.Chem., 56, 2013
|
|
