8HVP
 
 | | STRUCTURE AT 2.5-ANGSTROMS RESOLUTION OF CHEMICALLY SYNTHESIZED HUMAN IMMUNODEFICIENCY VIRUS TYPE 1 PROTEASE COMPLEXED WITH A HYDROXYETHYLENE*-BASED INHIBITOR | | Descriptor: | HIV-1 PROTEASE, INHIBITOR VAL-SER-GLN-ASN-LEU-PSI(CH(OH)-CH2)-VAL-ILE-VAL (U-85548E) | | Authors: | Jaskolski, M, Miller, M, Tomasselli, A.G, Sawyer, T.K, Staples, D.G, Heinrikson, R.L, Schneider, J, Kent, S.B.H, Wlodawer, A. | | Deposit date: | 1990-10-26 | | Release date: | 1993-10-31 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure at 2.5-A resolution of chemically synthesized human immunodeficiency virus type 1 protease complexed with a hydroxyethylene-based inhibitor.
Biochemistry, 30, 1991
|
|
1PPA
 
 | | THE CRYSTAL STRUCTURE OF A LYSINE 49 PHOSPHOLIPASE A2 FROM THE VENOM OF THE COTTONMOUTH SNAKE AT 2.0 ANGSTROMS RESOLUTION | | Descriptor: | ANILINE, PHOSPHOLIPASE A2 | | Authors: | Holland, D.R, Clancy, L.L, Muchmore, S.W, Rydel, T.J, Einspahr, H.M, Finzel, B.C, Heinrikson, R.L, Watenpaugh, K.D. | | Deposit date: | 1991-10-29 | | Release date: | 1994-01-31 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The crystal structure of a lysine 49 phospholipase A2 from the venom of the cottonmouth snake at 2.0-A resolution.
J.Biol.Chem., 265, 1990
|
|
1QTN
 
 | |
3KJF
 
 | | Caspase 3 Bound to a covalent inhibitor | | Descriptor: | (3S)-3-({[(5S,10aS)-2-{(2S)-4-carboxy-2-[(phenylacetyl)amino]butyl}-1,3-dioxo-2,3,5,7,8,9,10,10a-octahydro-1H-[1,2,4]triazolo[1,2-a]cinnolin-5-yl]carbonyl}amino)-4-oxopentanoic acid, Caspase-3 | | Authors: | Kamtekar, S, Watt, W, Finzel, B.C, Harris, M.S, Blinn, J, Wang, Z, Tomasselli, A.G. | | Deposit date: | 2009-11-03 | | Release date: | 2010-08-11 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Kinetic and structural characterization of caspase-3 and caspase-8 inhibition by a novel class of irreversible inhibitors.
Biochim.Biophys.Acta, 1804, 2010
|
|
3KJN
 
 | | Caspase 8 bound to a covalent inhibitor | | Descriptor: | (3S)-3-({[(5S)-2-{2-[(1H-benzimidazol-5-ylcarbonyl)amino]ethyl}-7-(cyclohexylmethyl)-1,3-dioxo-2,3,5,8-tetrahydro-1H-[1,2,4]triazolo[1,2-a]pyridazin-5-yl]carbonyl}amino)-4-oxopentanoic acid, 2,3-DIHYDROXY-1,4-DITHIOBUTANE, Caspase-8 | | Authors: | Kamtekar, S, Watt, W, Finzel, B.C, Harris, M.S, Blinn, J, Wang, Z, Tomasselli, A.G. | | Deposit date: | 2009-11-03 | | Release date: | 2010-08-11 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Kinetic and structural characterization of caspase-3 and caspase-8 inhibition by a novel class of irreversible inhibitors.
Biochim.Biophys.Acta, 1804, 2010
|
|
3KJQ
 
 | | Caspase 8 with covalent inhibitor | | Descriptor: | (3S)-3-({[(5S,8R)-2-(3-carboxypropyl)-8-(2-{[(4-chlorophenyl)acetyl]amino}ethyl)-1,3-dioxo-2,3,5,8-tetrahydro-1H-[1,2,4]triazolo[1,2-a]pyridazin-5-yl]carbonyl}amino)-4-oxopentanoic acid, Caspase-8 | | Authors: | Kamtekar, S, Watt, W, Finzel, B.C, Harris, M.S, Blinn, J, Wang, Z, Tomasselli, A.G. | | Deposit date: | 2009-11-03 | | Release date: | 2010-08-11 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Kinetic and structural characterization of caspase-3 and caspase-8 inhibition by a novel class of irreversible inhibitors.
Biochim.Biophys.Acta, 1804, 2010
|
|
1BHQ
 
 | | MAC-1 I DOMAIN CADMIUM COMPLEX | | Descriptor: | ACETYL GROUP, CADMIUM ION, CD11B | | Authors: | Baldwin, E.T. | | Deposit date: | 1998-06-10 | | Release date: | 1998-11-04 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Cation binding to the integrin CD11b I domain and activation model assessment
Structure, 6, 1998
|
|
1BHO
 
 | | MAC-1 I DOMAIN MAGNESIUM COMPLEX | | Descriptor: | CD11B, MAGNESIUM ION | | Authors: | Baldwin, E.T. | | Deposit date: | 1998-06-10 | | Release date: | 1998-11-04 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Cation binding to the integrin CD11b I domain and activation model assessment
Structure, 6, 1998
|
|
1GNN
 
 | | HIV-1 PROTEASE MUTANT WITH VAL 82 REPLACED BY ASN (V82N) COMPLEXED WITH U89360E (INHIBITOR) | | Descriptor: | HIV-1 PROTEASE, N-[[1-[N-ACETAMIDYL]-[1-CYCLOHEXYLMETHYL-2-HYDROXY-4-ISOPROPYL]-BUT-4-YL]-CARBONYL]-GLUTAMINYL-ARGINYL-AMIDE | | Authors: | Hong, L, Treharne, A, Hartsuck, J.A, Foundling, S, Tang, J. | | Deposit date: | 1996-05-04 | | Release date: | 1996-11-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structures of complexes of a peptidic inhibitor with wild-type and two mutant HIV-1 proteases.
Biochemistry, 35, 1996
|
|
7HVP
 
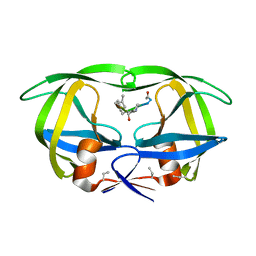 | | X-RAY CRYSTALLOGRAPHIC STRUCTURE OF A COMPLEX BETWEEN A SYNTHETIC PROTEASE OF HUMAN IMMUNODEFICIENCY VIRUS 1 AND A SUBSTRATE-BASED HYDROXYETHYLAMINE INHIBITOR | | Descriptor: | HIV-1 PROTEASE, INHIBITOR ACE-SER-LEU-ASN-PHE-PSI(CH(OH)-CH2N)-PRO-ILE VME (JG-365) | | Authors: | Swain, A.L, Miller, M.M, Green, J, Rich, D.H, Schneider, J, Kent, S.B.H, Wlodawer, A. | | Deposit date: | 1990-09-13 | | Release date: | 1993-07-15 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | X-ray crystallographic structure of a complex between a synthetic protease of human immunodeficiency virus 1 and a substrate-based hydroxyethylamine inhibitor.
Proc.Natl.Acad.Sci.USA, 87, 1990
|
|
1VIB
 
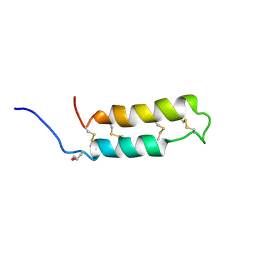 | | NMR SOLUTION STRUCTURE OF THE NEUROTOXIN B-IV, 20 STRUCTURES | | Descriptor: | NEUROTOXIN B-IV | | Authors: | Barnham, K.J, Dyke, T.R, Kem, W.R, Norton, R.S. | | Deposit date: | 1996-11-25 | | Release date: | 1997-05-15 | | Last modified: | 2017-11-29 | | Method: | SOLUTION NMR | | Cite: | Structure of neurotoxin B-IV from the marine worm Cerebratulus lacteus: a helical hairpin cross-linked by disulphide bonding.
J.Mol.Biol., 268, 1997
|
|
2ORA
 
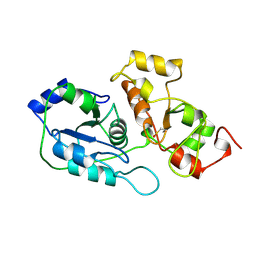 | | RHODANESE (THIOSULFATE: CYANIDE SULFURTRANSFERASE) | | Descriptor: | OXIDIZED RHODANESE | | Authors: | Gliubich, F, Gazerro, M, Zanotti, G, Delbono, S, Berni, R. | | Deposit date: | 1996-02-22 | | Release date: | 1996-08-01 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Active site structural features for chemically modified forms of rhodanese.
J.Biol.Chem., 271, 1996
|
|
1IDN
 
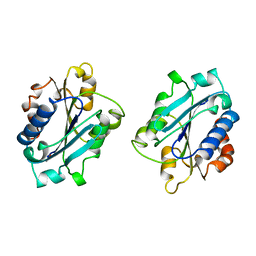 | | MAC-1 I DOMAIN METAL FREE | | Descriptor: | CD11B | | Authors: | Baldwin, E.T. | | Deposit date: | 1998-06-10 | | Release date: | 1998-11-04 | | Last modified: | 2019-07-17 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Cation binding to the integrin CD11b I domain and activation model assessment
Structure, 6, 1998
|
|
1I1B
 
 | |
4FM8
 
 | | Crystal Structure of BACE with Compound 12a | | Descriptor: | (5R,7S)-1-(3-fluorophenyl)-3,7-dimethyl-8-[3-(propan-2-yloxy)benzyl]-2-thia-1,3,8-triazaspiro[4.5]decane 2,2-dioxide, 1,2-ETHANEDIOL, Beta-secretase 1, ... | | Authors: | Vajdos, F.F, Varghese, A.H. | | Deposit date: | 2012-06-15 | | Release date: | 2012-10-03 | | Last modified: | 2013-06-19 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Spirocyclic sulfamides as beta-secretase 1 (BACE-1) inhibitors for the treatment of Alzheimer's disease: utilization of structure based drug design, WaterMap, and CNS penetration studies to identify centrally efficacious inhibitors.
J.Med.Chem., 55, 2012
|
|
4FM7
 
 | | Crystal Structure of BACE with Compound 14g | | Descriptor: | 4-{[(5R,7S)-1-(3-fluorophenyl)-3,7-dimethyl-2,2-dioxido-2-thia-1,3,8-triazaspiro[4.5]dec-8-yl]methyl}-2-(propan-2-yloxy)phenol, Beta-secretase 1, ZINC ION | | Authors: | Vajdos, F.F, Varghese, A.H. | | Deposit date: | 2012-06-15 | | Release date: | 2012-10-03 | | Last modified: | 2013-06-19 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Spirocyclic sulfamides as beta-secretase 1 (BACE-1) inhibitors for the treatment of Alzheimer's disease: utilization of structure based drug design, WaterMap, and CNS penetration studies to identify centrally efficacious inhibitors.
J.Med.Chem., 55, 2012
|
|
1HIV
 
 | | CRYSTAL STRUCTURE OF A COMPLEX OF HIV-1 PROTEASE WITH A DIHYDROETHYLENE-CONTAINING INHIBITOR: COMPARISONS WITH MOLECULAR MODELING | | Descriptor: | 4-[(2R)-3-{[(1S,2S,3R,4S)-1-(cyclohexylmethyl)-2,3-dihydroxy-5-methyl-4-({(1S,2R)-2-methyl-1-[(pyridin-2-ylmethyl)carbamoyl]butyl}carbamoyl)hexyl]amino}-2-{[(naphthalen-1-yloxy)acetyl]amino}-3-oxopropyl]-1H-imidazol-3-ium, HIV-1 PROTEASE | | Authors: | Thanki, N, Wlodawer, A. | | Deposit date: | 1992-02-12 | | Release date: | 1993-10-31 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of a complex of HIV-1 protease with a dihydroxyethylene-containing inhibitor: comparisons with molecular modeling.
Protein Sci., 1, 1992
|
|
1IVP
 
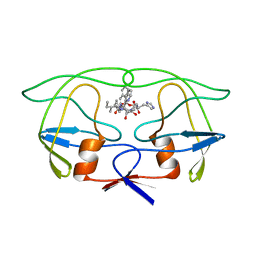 | | THE CRYSTALLOGRAPHIC STRUCTURE OF THE PROTEASE FROM HUMAN IMMUNODEFICIENCY VIRUS TYPE 2 WITH TWO SYNTHETIC PEPTIDIC TRANSITION STATE ANALOG INHIBITORS | | Descriptor: | 4-[(2R)-3-{[(1S,2S,3R,4S)-1-(cyclohexylmethyl)-2,3-dihydroxy-5-methyl-4-({(1S,2R)-2-methyl-1-[(pyridin-2-ylmethyl)carba moyl]butyl}carbamoyl)hexyl]amino}-2-{[(naphthalen-1-yloxy)acetyl]amino}-3-oxopropyl]-1H-imidazol-3-ium, HIV-2 PROTEASE | | Authors: | Mulichak, A.M, Watenpaugh, K.D. | | Deposit date: | 1993-03-18 | | Release date: | 1993-07-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The crystallographic structure of the protease from human immunodeficiency virus type 2 with two synthetic peptidic transition state analog inhibitors.
J.Biol.Chem., 268, 1993
|
|
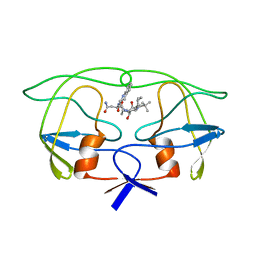
1IVQ
 
 | |
3UDH
 
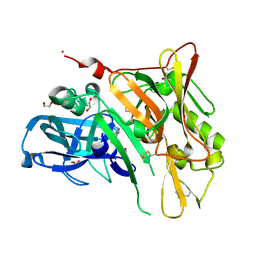 | | Crystal Structure of BACE with Compound 1 | | Descriptor: | (3S)-spiro[indole-3,3'-pyrrolidin]-2(1H)-one, 1,2-ETHANEDIOL, Beta-secretase 1, ... | | Authors: | Efremov, I.V, Vajdos, F.F, Borzilleri, K, Capetta, S, Dorff, P, Dutra, J, Mansour, M, Oborski, C, O'Connell, T, O'Sullivan, T.J, Pandit, J, Wang, H, Withka, J. | | Deposit date: | 2011-10-28 | | Release date: | 2012-04-18 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Discovery and optimization of a novel spiropyrrolidine inhibitor of {beta}-secretase (BACE1) through fragment-based drug design.
J.Med.Chem., 55, 2012
|
|
3UDQ
 
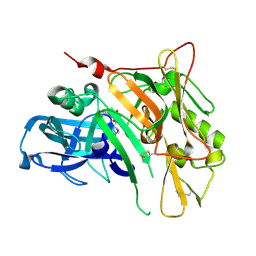 | | Crystal Structure of BACE with Compound 13 | | Descriptor: | (4S)-6-bromo-1,1-dioxido-3,4-dihydro-2H-thiochromen-4-yl (3S,5'R)-2-oxo-1,2-dihydrospiro[indole-3,3'-pyrrolidine]-5'-carboxylate, Beta-secretase 1 | | Authors: | Efremov, I.V, Vajdos, F.F, Borzilleri, K, Capetta, S, Dorff, P, Dutra, J, Mansour, M, Oborski, C, O'Connell, T, O'Sullivan, T.J, Pandit, J, Wang, H, Withka, J. | | Deposit date: | 2011-10-28 | | Release date: | 2012-04-18 | | Last modified: | 2018-04-04 | | Method: | X-RAY DIFFRACTION (2.73 Å) | | Cite: | Discovery and optimization of a novel spiropyrrolidine inhibitor of {beta}-secretase (BACE1) through fragment-based drug design.
J.Med.Chem., 55, 2012
|
|
3UDM
 
 | | Crystal Structure of BACE with Compound 8 | | Descriptor: | 1,2-ETHANEDIOL, Beta-secretase 1, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Efremov, I.V, Vajdos, F.F, Borzilleri, K, Capetta, S, Dorff, P, Dutra, J, Mansour, M, Oborski, C, O'Connell, T, O'Sullivan, T.J, Pandit, J, Wang, H, Withka, J. | | Deposit date: | 2011-10-28 | | Release date: | 2012-04-18 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Discovery and optimization of a novel spiropyrrolidine inhibitor of {beta}-secretase (BACE1) through fragment-based drug design.
J.Med.Chem., 55, 2012
|
|
3UDJ
 
 | | Crystal Structure of BACE with Compound 5 | | Descriptor: | 1,2-ETHANEDIOL, Beta-secretase 1, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Efremov, I.V, Vajdos, F.F, Borzilleri, K, Capetta, S, Dorff, P, Dutra, J, Mansour, M, Oborski, C, O'Connell, T, O'Sullivan, T.J, Pandit, J, Wang, H, Withka, J. | | Deposit date: | 2011-10-28 | | Release date: | 2012-04-18 | | Last modified: | 2013-06-19 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Discovery and optimization of a novel spiropyrrolidine inhibitor of {beta}-secretase (BACE1) through fragment-based drug design.
J.Med.Chem., 55, 2012
|
|
3UDR
 
 | | Crystal Structure of BACE with Compound 14 | | Descriptor: | 1,2-ETHANEDIOL, 1-cyanocyclohexyl (3S,5'R)-2-oxo-1,2-dihydrospiro[indole-3,3'-pyrrolidine]-5'-carboxylate, Beta-secretase 1, ... | | Authors: | Efremov, I.V, Vajdos, F.F, Borzilleri, K, Capetta, S, Dorff, P, Dutra, J, Mansour, M, Oborski, C, O'Connell, T, O'Sullivan, T.J, Pandit, J, Wang, H, Withka, J. | | Deposit date: | 2011-10-28 | | Release date: | 2012-04-18 | | Last modified: | 2018-04-04 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Discovery and optimization of a novel spiropyrrolidine inhibitor of {beta}-secretase (BACE1) through fragment-based drug design.
J.Med.Chem., 55, 2012
|
|
3UDN
 
 | | Crystal Structure of BACE with Compound 9 | | Descriptor: | 1,2-ETHANEDIOL, 4-cyanobenzyl (3S,5'R)-2-oxo-1,2-dihydrospiro[indole-3,3'-pyrrolidine]-5'-carboxylate, Beta-secretase 1, ... | | Authors: | Efremov, I.V, Vajdos, F.F, Borzilleri, K, Capetta, S, Dorff, P, Dutra, J, Mansour, M, Oborski, C, O'Connell, T, O'Sullivan, T.J, Pandit, J, Wang, H, Withka, J. | | Deposit date: | 2011-10-28 | | Release date: | 2012-04-18 | | Last modified: | 2018-04-04 | | Method: | X-RAY DIFFRACTION (2.193 Å) | | Cite: | Discovery and optimization of a novel spiropyrrolidine inhibitor of {beta}-secretase (BACE1) through fragment-based drug design.
J.Med.Chem., 55, 2012
|
|
