1BOH
 
 | | SULFUR-SUBSTITUTED RHODANESE (ORTHORHOMBIC FORM) | | Descriptor: | RHODANESE | | Authors: | Gliubich, F, Berni, R, Cianci, M, Trevino, R.J, Horowitz, P.M, Zanotti, G. | | Deposit date: | 1998-08-04 | | Release date: | 1999-04-27 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | NH2-terminal sequence truncation decreases the stability of bovine rhodanese, minimally perturbs its crystal structure, and enhances interaction with GroEL under native conditions.
J.Biol.Chem., 274, 1999
|
|
1BOI
 
 | | N-TERMINALLY TRUNCATED RHODANESE | | Descriptor: | RHODANESE | | Authors: | Gliubich, F, Berni, R, Cianci, M, Trevino, R.J, Horowitz, P.M, Zanotti, G. | | Deposit date: | 1998-08-04 | | Release date: | 1999-04-27 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | NH2-terminal sequence truncation decreases the stability of bovine rhodanese, minimally perturbs its crystal structure, and enhances interaction with GroEL under native conditions.
J.Biol.Chem., 274, 1999
|
|
2ORA
 
 | | RHODANESE (THIOSULFATE: CYANIDE SULFURTRANSFERASE) | | Descriptor: | OXIDIZED RHODANESE | | Authors: | Gliubich, F, Gazerro, M, Zanotti, G, Delbono, S, Berni, R. | | Deposit date: | 1996-02-22 | | Release date: | 1996-08-01 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Active site structural features for chemically modified forms of rhodanese.
J.Biol.Chem., 271, 1996
|
|
1ORB
 
 | | ACTIVE SITE STRUCTURAL FEATURES FOR CHEMICALLY MODIFIED FORMS OF RHODANESE | | Descriptor: | ACETATE ION, CARBOXYMETHYLATED RHODANESE | | Authors: | Gliubich, F, Gazerro, M, Zanotti, G, Delbono, S, Berni, R. | | Deposit date: | 1995-07-24 | | Release date: | 1995-10-15 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Active site structural features for chemically modified forms of rhodanese.
J.Biol.Chem., 271, 1996
|
|
1AOW
 
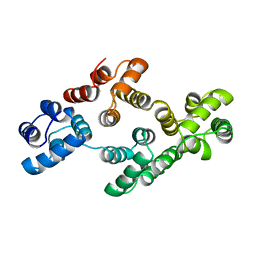 | | ANNEXIN IV | | Descriptor: | ANNEXIN IV | | Authors: | Zanotti, G, Malpeli, G, Gliubich, F, Folli, C, Stoppini, M, Olivi, L, Savoia, A, Berni, R. | | Deposit date: | 1997-07-11 | | Release date: | 1998-01-14 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of the trigonal crystal form of bovine annexin IV.
Biochem.J., 329, 1998
|
|
1RHS
 
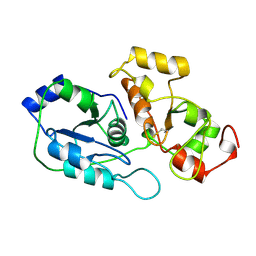 | | SULFUR-SUBSTITUTED RHODANESE | | Descriptor: | SULFUR-SUBSTITUTED RHODANESE | | Authors: | Zanotti, G, Gliubich, F, Colapietro, M, Barba, L. | | Deposit date: | 1997-07-16 | | Release date: | 1998-01-21 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | Structure of sulfur-substituted rhodanese at 1.36 A resolution.
Acta Crystallogr.,Sect.D, 54, 1998
|
|
3T36
 
 | |
3BSZ
 
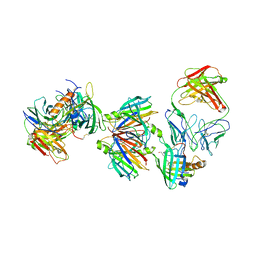 | | Crystal structure of the transthyretin-retinol binding protein-Fab complex | | Descriptor: | Fab fragment heavy chain, Fab fragment light chain, Plasma retinol-binding protein, ... | | Authors: | Zanotti, G, Cendron, L, Gliubich, F, Folli, C, Berni, R. | | Deposit date: | 2007-12-27 | | Release date: | 2008-11-11 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.38 Å) | | Cite: | Structural and mutational analyses of protein-protein interactions between transthyretin and retinol-binding protein.
Febs J., 275, 2008
|
|
4HJZ
 
 | | 1.9 A Crystal structure of E. coli MltE-E64Q with bound chitopentaose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ... | | Authors: | Fibriansah, G, Gliubich, F.I, Thunnissen, A.-M.W.H. | | Deposit date: | 2012-10-14 | | Release date: | 2012-10-24 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | On the Mechanism of Peptidoglycan Binding and Cleavage by the endo-Specific Lytic Transglycosylase MltE from Escherichia coli.
Biochemistry, 51, 2012
|
|
4HJY
 
 | | 2.4 A Crystal structure of E. coli MltE-E64Q with bound chitopentaose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Endo-type membrane-bound lytic murein transglycosylase A | | Authors: | Fibriansah, G, Gliubich, F.I, Thunnissen, A.-M.W.H. | | Deposit date: | 2012-10-14 | | Release date: | 2012-10-24 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | On the Mechanism of Peptidoglycan Binding and Cleavage by the endo-Specific Lytic Transglycosylase MltE from Escherichia coli.
Biochemistry, 51, 2012
|
|
4HJV
 
 | | Crystal structure of E. coli MltE with bound bulgecin and murodipeptide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-N-acetyl-beta-muramic acid, 4-O-(4-O-SULFONYL-N-ACETYLGLUCOSAMININYL)-5-METHYLHYDROXY-L-PROLINE-TAURINE, Endo-type membrane-bound lytic murein transglycosylase A, ... | | Authors: | Fibriansah, G, Gliubich, F.I, Thunnissen, A.-M.W.H. | | Deposit date: | 2012-10-14 | | Release date: | 2012-10-24 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | On the Mechanism of Peptidoglycan Binding and Cleavage by the endo-Specific Lytic Transglycosylase MltE from Escherichia coli.
Biochemistry, 51, 2012
|
|
3BT0
 
 | | Crystal structure of transthyretin variant V20S | | Descriptor: | Transthyretin | | Authors: | Zanotti, G, Folli, C, Cendron, L, Gliubich, F, Negro, A, Berni, R. | | Deposit date: | 2007-12-27 | | Release date: | 2008-11-11 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Structural and mutational analyses of protein-protein interactions between transthyretin and retinol-binding protein.
Febs J., 275, 2008
|
|
3FE5
 
 | | Crystal structure of 3-hydroxyanthranilate 3,4-dioxygenase from bovine kidney | | Descriptor: | 3-hydroxyanthranilate 3,4-dioxygenase, FE (III) ION | | Authors: | Dilovic, I, Gliubich, F, Malpeli, G, Zanotti, G, Matkovic-Calogovic, D. | | Deposit date: | 2008-11-27 | | Release date: | 2009-06-09 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Crystal structure of bovine 3-hydroxyanthranilate 3,4-dioxygenase.
Biopolymers, 2009
|
|
