4UIB
 
 | | Crystal structure of 3p in complex with tafCPB | | Descriptor: | (2S)-6-AMINO-2-[[(1R)-1-(CYCLOHEXYLMETHYL)-2-OXO-2-[[(2S)-1,7,7-TRIMETHYLNORBORNAN-2-YL]AMINO]ETHYL]ARBAMOYLAMINO]HEXANOIC ACID, ACETATE ION, CARBOXYPEPTIDASE B, ... | | Authors: | Halland, N, Broenstrup, M, Czech, J, Czechtizky, W, Evers, A, Follmann, M, Kohlmann, M, Schiell, M, Kurz, M, Schreuder, H.A, Kallus, C. | | Deposit date: | 2015-03-27 | | Release date: | 2015-06-03 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Novel Small Molecule Inhibitors of Activated Thrombin Activatable Fibrinolysis Inhibitor (Tafia) from Natural Product Anabaenopeptin.
J.Med.Chem., 58, 2015
|
|
4UIA
 
 | | Crystal structure of 3a in complex with tafCPB | | Descriptor: | (2S)-6-azanyl-2-[[(2R)-3-cyclohexyl-1-(3-methylbutylamino)-1-oxidanylidene-propan-2-yl]carbamoylamino]hexanoic acid, CARBOXYPEPTIDASE B, ZINC ION | | Authors: | Halland, N, Broenstrup, M, Czech, J, Czechtizky, W, Evers, A, Follmann, M, Kohlmann, M, Schiell, M, Kurz, M, Schreuder, H.A, Kallus, C. | | Deposit date: | 2015-03-27 | | Release date: | 2015-06-03 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Novel Small Molecule Inhibitors of Activated Thrombin Activatable Fibrinolysis Inhibitor (Tafia) from Natural Product Anabaenopeptin.
J.Med.Chem., 58, 2015
|
|
7PUE
 
 | | Human serum and glucocorticoid-regulated kinase 1 in complex with pyrazolopyridine inhibitor 3a | | Descriptor: | 6-[4-[[2,3-bis(chloranyl)phenyl]sulfonylamino]phenyl]-~{N}-[(3~{R})-pyrrolidin-3-yl]-2~{H}-pyrazolo[3,4-b]pyridine-4-carboxamide, GLYCEROL, Serine/threonine-protein kinase Sgk1 | | Authors: | Dreyer, M.K, Halland, N, Nazare, M. | | Deposit date: | 2021-09-29 | | Release date: | 2021-12-01 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.506 Å) | | Cite: | Rational Design of Highly Potent, Selective, and Bioavailable SGK1 Protein Kinase Inhibitors for the Treatment of Osteoarthritis.
J.Med.Chem., 65, 2022
|
|
3MGL
 
 | |
6JQN
 
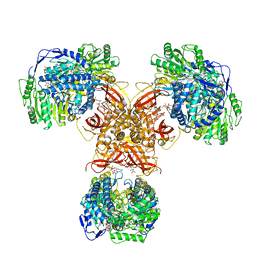 | | Structure of PaaZ, a bifunctional enzyme in complex with NADP+ and OCoA | | Descriptor: | Bifunctional protein PaaZ, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, OCTANOYL-COENZYME A | | Authors: | Gakher, L, Vinothkumar, K.R, Katagihallimath, N, Sowdhamini, R, Sathyanarayanan, N, Cannone, G. | | Deposit date: | 2019-03-31 | | Release date: | 2019-09-11 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Molecular basis for metabolite channeling in a ring opening enzyme of the phenylacetate degradation pathway.
Nat Commun, 10, 2019
|
|
6JQO
 
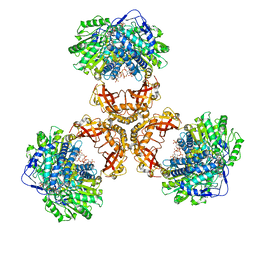 | | Structure of PaaZ, a bifunctional enzyme in complex with NADP+ and CCoA | | Descriptor: | Bifunctional protein PaaZ, CROTONYL COENZYME A, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Gakher, L, Vinothkumar, K.R, Katagihallimath, N, Sowdhamini, R, Sathyanarayanan, N, Cannone, G. | | Deposit date: | 2019-03-31 | | Release date: | 2019-09-11 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Molecular basis for metabolite channeling in a ring opening enzyme of the phenylacetate degradation pathway.
Nat Commun, 10, 2019
|
|
6JQL
 
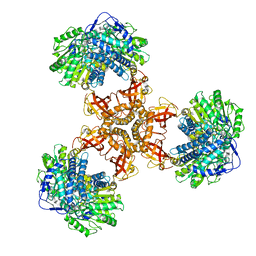 | | Structure of PaaZ, a bifunctional enzyme | | Descriptor: | Bifunctional protein PaaZ | | Authors: | Gakher, L, Vinothkumar, K.R, Katagihallimath, N, Sowdhamini, R, Sathyanarayanan, N, Cannone, G. | | Deposit date: | 2019-03-31 | | Release date: | 2019-09-11 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Molecular basis for metabolite channeling in a ring opening enzyme of the phenylacetate degradation pathway.
Nat Commun, 10, 2019
|
|
6JQM
 
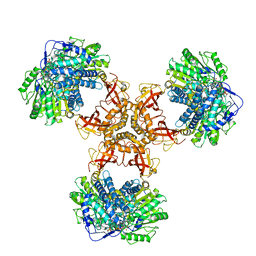 | | Structure of PaaZ with NADPH | | Descriptor: | Bifunctional protein PaaZ, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Gakher, L, Vinothkumar, K.R, Katagihallimath, N, Sowdhamini, R, Sathyanarayanan, N, Cannone, G. | | Deposit date: | 2019-03-31 | | Release date: | 2019-09-11 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Molecular basis for metabolite channeling in a ring opening enzyme of the phenylacetate degradation pathway.
Nat Commun, 10, 2019
|
|
5V43
 
 | | Engineered human IgG Fc domain aglyco801 | | Descriptor: | Ig gamma-1 chain C region | | Authors: | Yan, W, Marshall, N, Zhang, Y.J. | | Deposit date: | 2017-03-08 | | Release date: | 2017-06-21 | | Last modified: | 2017-09-27 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | IgG Fc domains that bind C1q but not effector Fc gamma receptors delineate the importance of complement-mediated effector functions.
Nat. Immunol., 18, 2017
|
|
5V4E
 
 | | Engineered human IgG Fc domain glyco801 (Fc801) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-2)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Yan, W, Marshall, N, Zhang, Y.J. | | Deposit date: | 2017-03-09 | | Release date: | 2017-06-21 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.216 Å) | | Cite: | IgG Fc domains that bind C1q but not effector Fc gamma receptors delineate the importance of complement-mediated effector functions.
Nat. Immunol., 18, 2017
|
|
2L5J
 
 | | structure of the spliceosomal phosphopeptide P140 (phosphorylated form) | | Descriptor: | U1 small nuclear ribonucleoprotein 70 kDa | | Authors: | Quinternet, M, Page, N, Schall, N, Strub, J, Chaloin, O, Decossas, M, Cung, M, van Dorsselaer, A, Briand, J, Muller, S. | | Deposit date: | 2010-11-02 | | Release date: | 2010-12-01 | | Last modified: | 2011-07-13 | | Method: | SOLUTION NMR | | Cite: | The spliceosomal phosphopeptide P140 controls the lupus disease by interacting with the HSC70 protein and via a mechanism mediated by gammadelta T cells.
Plos One, 4, 2009
|
|
2L5I
 
 | | structure of the spliceosomal phosphopeptide P140 (non-phosphorylated form) | | Descriptor: | U1 small nuclear ribonucleoprotein 70 kDa | | Authors: | Quinternet, M, Page, N, Schall, N, Strub, J, Chaloin, O, Decossas, M, Cung, M, van Dorsselaer, A, Briand, J, Muller, S. | | Deposit date: | 2010-11-02 | | Release date: | 2010-12-01 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | The spliceosomal phosphopeptide P140 controls the lupus disease by interacting with the HSC70 protein and via a mechanism mediated by gammadelta T cells.
Plos One, 4, 2009
|
|
1LQU
 
 | | Mycobacterium tuberculosis FprA in complex with NADPH | | Descriptor: | ACETATE ION, FLAVIN-ADENINE DINUCLEOTIDE, FprA, ... | | Authors: | Bossi, R.T, Aliverti, A, Raimondi, D, Fischer, F, Zanetti, G, Ferrari, D, Tahallah, N, Maier, C.S, Heck, A.J.R, Rizzi, M, Mattevi, A, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2002-05-13 | | Release date: | 2002-07-31 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | A covalent modification of NADP+ revealed by the atomic resolution structure of FprA, a Mycobacterium tuberculosis oxidoreductase.
Biochemistry, 41, 2002
|
|
1LQT
 
 | | A covalent modification of NADP+ revealed by the atomic resolution structure of FprA, a Mycobacterium tuberculosis oxidoreductase | | Descriptor: | 4-OXO-NICOTINAMIDE-ADENINE DINUCLEOTIDE PHOSPHATE, ACETATE ION, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Bossi, R.T, Aliverti, A, Raimondi, D, Fischer, F, Zanetti, G, Ferrari, D, Tahallah, N, Maier, C.S, Heck, A.J.R, Rizzi, M, Mattevi, A, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2002-05-13 | | Release date: | 2002-07-31 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | A covalent modification of NADP+ revealed by the atomic resolution structure of FprA, a Mycobacterium tuberculosis oxidoreductase.
Biochemistry, 41, 2002
|
|
5DA8
 
 | | Crystal structure of chaperonin GroEL from | | Descriptor: | 60 kDa chaperonin, CALCIUM ION, MAGNESIUM ION, ... | | Authors: | Chang, C, Marshall, N, Feldmann, B, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2015-08-19 | | Release date: | 2015-10-14 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of chaperonin GroEL from
To Be Published
|
|
8CXK
 
 | | Structure of the C. elegans HIM-3 R93Y mutant | | Descriptor: | HORMA domain-containing protein | | Authors: | Ego, K.M, Russo, A, Giacopazzi, S, Deshong, A, Menon, M, Ortiz, V, Bhalla, N, Corbett, K.D. | | Deposit date: | 2022-05-21 | | Release date: | 2023-05-17 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | The conserved AAA ATPase PCH-2 distributes its regulation of meiotic prophase events through multiple meiotic HORMADs in C. elegans.
Plos Genet., 19, 2023
|
|
3ZLS
 
 | | Crystal structure of MEK1 in complex with fragment 6 | | Descriptor: | 1H-PYRROLO[2,3-B]PYRIDINE-3-CARBOXYLIC ACID, DUAL SPECIFICITY MITOGEN-ACTIVATED PROTEIN KINASE KINASE 1, SODIUM ION | | Authors: | Amaning, K, Lowinsky, M, Vallee, F, Steier, V, Marcireau, C, Ugolini, A, Delorme, C, McCort, G, Andouche, C, Vougier, S, Llopart, S, Halland, N, Rak, A. | | Deposit date: | 2013-02-04 | | Release date: | 2013-05-22 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The Use of Virtual Screening and Differential Scanning Fluorimetry for the Rapid Identification of Fragments Active Against Mek1.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
3ZM4
 
 | | Crystal structure of MEK1 in complex with fragment 1 | | Descriptor: | 7-chloranyl-6-[(3S)-pyrrolidin-3-yl]oxy-2H-isoquinolin-1-one, DUAL SPECIFICITY MITOGEN-ACTIVATED PROTEIN KINASE KINASE 1 | | Authors: | Amaning, K, Lowinsky, M, Vallee, F, Steier, V, Marcireau, C, Ugolini, A, Delorme, C, McCort, G, Andouche, C, Vougier, S, Llopart, S, Halland, N, Rak, A. | | Deposit date: | 2013-02-05 | | Release date: | 2013-05-22 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | The Use of Virtual Screening and Differential Scanning Fluorimetry for the Rapid Identification of Fragments Active Against Mek1.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
3ZLW
 
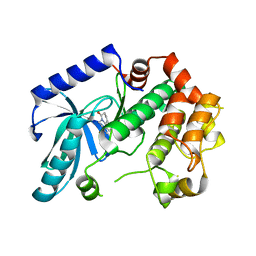 | | Crystal structure of MEK1 in complex with fragment 3 | | Descriptor: | (1R)-1-hydroxy-1-methyl-2,3,6,7-tetrahydro-1H,5H-pyrido[3,2,1-ij]quinolin-5-one, DUAL SPECIFICITY MITOGEN-ACTIVATED PROTEIN KINASE KINASE 1 MAPK/ERK KINASE 1, MEK 1, ... | | Authors: | Amaning, K, Lowinsky, M, Vallee, F, Steier, V, Marcireau, C, Ugolini, A, Delorme, C, McCort, G, Andouche, C, Vougier, S, Llopart, S, Halland, N, Rak, A. | | Deposit date: | 2013-02-04 | | Release date: | 2013-05-22 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | The Use of Virtual Screening and Differential Scanning Fluorimetry for the Rapid Identification of Fragments Active Against Mek1.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
3ZLX
 
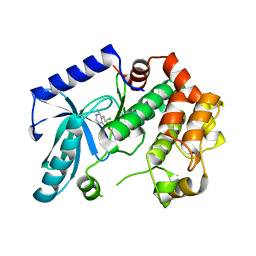 | | Crystal structure of MEK1 in complex with fragment 18 | | Descriptor: | 7-choro-6-[(3R)-pyrrolidin-3-ylmethoxy]isoquinolin-1(2H)-one, DUAL SPECIFICITY MITOGEN-ACTIVATED PROTEIN KINASE KINASE 1 | | Authors: | Amaning, K, Lowinsky, M, Vallee, F, Steier, V, Marcireau, C, Ugolini, A, Delorme, C, McCort, G, Andouche, C, Vougier, S, Llopart, S, Halland, N, Rak, A. | | Deposit date: | 2013-02-04 | | Release date: | 2013-05-22 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The Use of Virtual Screening and Differential Scanning Fluorimetry for the Rapid Identification of Fragments Active Against Mek1.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
3ZLY
 
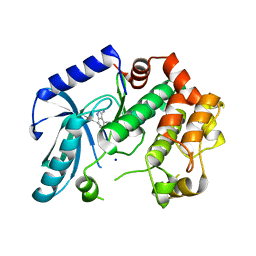 | | Crystal structure of MEK1 in complex with fragment 8 | | Descriptor: | 3-AMINO-1H-INDAZOLE-4-CARBONITRILE, DUAL SPECIFICITY MITOGEN-ACTIVATED PROTEIN KINASE KINASE 1, SODIUM ION | | Authors: | Amaning, K, Lowinsky, M, Vallee, F, Steier, V, Marcireau, C, Ugolini, A, Delorme, C, McCort, G, Andouche, C, Vougier, S, Llopart, S, Halland, N, Rak, A. | | Deposit date: | 2013-02-04 | | Release date: | 2013-05-22 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | The Use of Virtual Screening and Differential Scanning Fluorimetry for the Rapid Identification of Fragments Active Against Mek1.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
3M1G
 
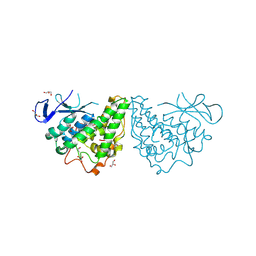 | | The structure of a putative glutathione S-transferase from Corynebacterium glutamicum | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, Putative glutathione S-transferase | | Authors: | Cuff, M.E, Marshall, N, Cobb, G, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-03-04 | | Release date: | 2010-04-14 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The structure of a putative glutathione S-transferase from Corynebacterium glutamicum
TO BE PUBLISHED
|
|
3PVZ
 
 | | UDP-N-acetylglucosamine 4,6-dehydratase from Vibrio fischeri | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SODIUM ION, THIOCYANATE ION, ... | | Authors: | Osipiuk, J, Marshall, N, Tesar, C, Pearson, L, Buck, K, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-12-07 | | Release date: | 2011-01-12 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | UDP-N-acetylglucosamine 4,6-dehydratase from Vibrio fischeri.
To be Published
|
|
3OIZ
 
 | | Crystal structure of antisigma-factor antagonist, STAS domain from Rhodobacter sphaeroides | | Descriptor: | Antisigma-factor antagonist, STAS | | Authors: | Chang, C, Marshall, N, Freeman, L, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-08-20 | | Release date: | 2010-09-08 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structure of antisigma-factor antagonist, STAS domain from Rhodobacter sphaeroides
To be Published
|
|
3U2Z
 
 | | Activator-Bound Structure of Human Pyruvate Kinase M2 | | Descriptor: | 1,6-di-O-phosphono-beta-D-fructofuranose, 6-(3-aminobenzyl)-4-methyl-2-methylsulfinyl-4,6-dihydro-5H-thieno[2',3':4,5]pyrrolo[2,3-d]pyridazin-5-one, Pyruvate kinase isozymes M1/M2, ... | | Authors: | Hong, B, Dimov, S, Tempel, W, Auld, D, Thomas, C, Boxer, M, Jianq, J.-K, Skoumbourdis, A, Min, S, Southall, N, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Weigelt, J, Inglese, J, Park, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2011-10-04 | | Release date: | 2012-09-12 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Pyruvate kinase M2 activators promote tetramer formation and suppress tumorigenesis.
Nat.Chem.Biol., 8, 2012
|
|
