7KKL
 
 | | SARS-CoV-2 Spike in complex with neutralizing nanobody mNb6 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Schoof, M.S, Faust, B.F, Saunders, R.A, Sangwan, S, Rezelj, V, Hoppe, N, Boone, M, Billesboelle, C.B, Puchades, C, Azumaya, C.M, Kratochvil, H.T, Zimanyi, M, Desphande, I, Liang, J, Dickinson, S, Nguyen, H.C, Chio, C.M, Merz, G.E, Thompson, M.C, Diwanji, D, Schaefer, K, Anand, A.A, Dobzinski, N, Zha, B.S, Simoneau, C.R, Leon, K, White, K.M, Chio, U.S, Gupta, M, Jin, M, Li, F, Liu, Y, Zhang, K, Bulkley, D, Sun, M, Smith, A.M, Rizo, A.N, Moss, F, Brilot, A.F, Pourmal, S, Trenker, R, Pospiech, T, Gupta, S, Barsi-Rhyne, B, Belyy, V, Barile-Hill, A.W, Nock, S, Liu, Y, Krogan, N.J, Ralston, C.Y, Swaney, D.L, Garcia-Sastre, A, Ott, M, Vignuzzi, M, Walter, P, Manglik, A, QCRG Structural Biology Consortium | | Deposit date: | 2020-10-27 | | Release date: | 2020-11-11 | | Last modified: | 2021-04-21 | | Method: | ELECTRON MICROSCOPY (2.85 Å) | | Cite: | An ultrapotent synthetic nanobody neutralizes SARS-CoV-2 by stabilizing inactive Spike.
Science, 370, 2020
|
|
7KKK
 
 | | SARS-CoV-2 Spike in complex with neutralizing nanobody Nb6 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Schoof, M.S, Faust, B.F, Saunders, R.A, Sangwan, S, Rezelj, V, Hoppe, N, Boone, M, Billesboelle, C.B, Puchades, C, Azumaya, C.M, Kratochvil, H.T, Zimanyi, M, Desphande, I, Liang, J, Dickinson, S, Nguyen, H.C, Chio, C.M, Merz, G.E, Thompson, M.C, Diwanji, D, Schaefer, K, Anand, A.A, Dobzinski, N, Zha, B.S, Simoneau, C.R, Leon, K, White, K.M, Chio, U.S, Gupta, M, Jin, M, Li, F, Liu, Y, Zhang, K, Bulkley, D, Sun, M, Smith, A.M, Rizo, A.N, Moss, F, Brilot, A.F, Pourmal, S, Trenker, R, Pospiech, T, Gupta, S, Barsi-Rhyne, B, Belyy, V, Barile-Hill, A.W, Nock, S, Liu, Y, Krogan, N.J, Ralston, C.Y, Swaney, D.L, Garcia-Sastre, A, Ott, M, Vignuzzi, M, Walter, P, Manglik, A, QCRG Structural Biology Consortium | | Deposit date: | 2020-10-27 | | Release date: | 2020-11-11 | | Last modified: | 2021-04-21 | | Method: | ELECTRON MICROSCOPY (3.03 Å) | | Cite: | An ultrapotent synthetic nanobody neutralizes SARS-CoV-2 by stabilizing inactive Spike.
Science, 370, 2020
|
|
2PI0
 
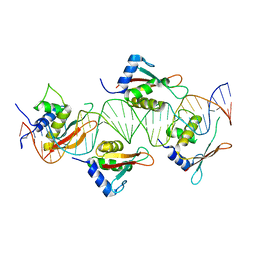 | | Crystal Structure of IRF-3 bound to the PRDIII-I regulatory element of the human interferon-B enhancer | | Descriptor: | Interferon regulatory factor 3, PRDIII-I region of human interferon-B promoter strand 1, PRDIII-I region of human interferon-B promoter strand 2 | | Authors: | Escalante, C.R, Nistal-Villan, E, Leyi, S, Garcia-Sastre, A, Aggarwal, A.K. | | Deposit date: | 2007-04-12 | | Release date: | 2007-10-30 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Structure of IRF-3 bound to the PRDIII-I regulatory element of the human interferon-beta enhancer.
Mol.Cell, 26, 2007
|
|
3PSE
 
 | | Structure of a viral OTU domain protease bound to interferon-stimulated gene 15 (ISG15) | | Descriptor: | 1.7.6 3-bromanylpropan-1-amine, GLYCEROL, RNA polymerase, ... | | Authors: | Bacik, J.P, James, T.W, Frias-Staheli, N, Garcia-Sastre, A, Mark, B.L. | | Deposit date: | 2010-12-01 | | Release date: | 2011-01-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for the removal of ubiquitin and interferon-stimulated gene 15 by a viral ovarian tumor domain-containing protease.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3PT2
 
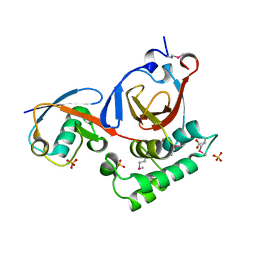 | | Structure of a viral OTU domain protease bound to Ubiquitin | | Descriptor: | 1.7.6 3-bromanylpropan-1-amine, ACETATE ION, RNA polymerase, ... | | Authors: | James, T.W, Bacik, J.P, Frias-Staheli, N, Garcia-Sastre, A, Mark, B.L. | | Deposit date: | 2010-12-02 | | Release date: | 2011-01-19 | | Last modified: | 2023-05-31 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for the removal of ubiquitin and interferon-stimulated gene 15 by a viral ovarian tumor domain-containing protease.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
7KKJ
 
 | | Structure of anti-SARS-CoV-2 Spike nanobody mNb6 | | Descriptor: | CHLORIDE ION, SULFATE ION, Synthetic nanobody mNb6 | | Authors: | Schoof, M.S, Faust, B.F, Saunders, R.A, Sangwan, S, Rezelj, V, Hoppe, N, Boone, M, Billesboelle, C.B, Puchades, C, Azumaya, C.M, Kratochvil, H.T, Zimanyi, M, Desphande, I, Liang, J, Dickinson, S, Nguyen, H.C, Chio, C.M, Merz, G.E, Thompson, M.C, Diwanji, D, Schaefer, K, Anand, A.A, Dobzinski, N, Zha, B.S, Simoneau, C.R, Leon, K, White, K.M, Chio, U.S, Gupta, M, Jin, M, Li, F, Liu, Y, Zhang, K, Bulkley, D, Sun, M, Smith, A.M, Rizo, A.N, Moss, F, Brilot, A.F, Pourmal, S, Trenker, R, Pospiech, T, Gupta, S, Barsi-Rhyne, B, Belyy, V, Barile-Hill, A.W, Nock, S, Liu, Y, Krogan, N.J, Ralston, C.Y, Swaney, D.L, Garcia-Sastre, A, Ott, M, Vignuzzi, M, Walter, P, Manglik, A, QCRG Structural Biology Consortium | | Deposit date: | 2020-10-27 | | Release date: | 2020-11-25 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | An ultrapotent synthetic nanobody neutralizes SARS-CoV-2 by stabilizing inactive Spike.
Science, 370, 2020
|
|
5V2A
 
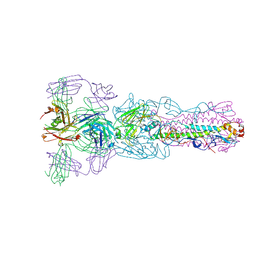 | | Crystal structure of Fab H7.167 in complex with influenza virus hemagglutinin from A/Shanghai/02/2013 (H7N9) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of H7.167 antibody, Hemagglutinin, ... | | Authors: | Zhang, H, Zhu, X, Wilson, I.A. | | Deposit date: | 2017-03-03 | | Release date: | 2017-04-05 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (4.656 Å) | | Cite: | H7N9 influenza virus neutralizing antibodies that possess few somatic mutations.
J. Clin. Invest., 126, 2016
|
|
8VCC
 
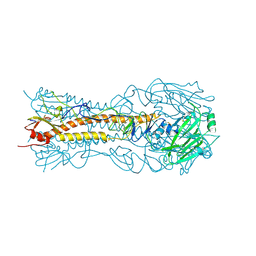 | |
6WCZ
 
 | | CryoEM structure of full-length ZIKV NS5-hSTAT2 complex | | Descriptor: | Non-structural protein 5, Signal transducer and activator of transcription 2, ZINC ION | | Authors: | Boxiao, W, Stephanie, T, Kang, Z, Maria, T.S, Jian, F, Jiuwei, L, Linfeng, G, Wendan, R, Yanxiang, C, Ethan, C.V, HeaJin, H, Matthew, J.E, Sean, E.O, Adolfo, G.S, Hong, Z, Rong, H, Jikui, S. | | Deposit date: | 2020-03-31 | | Release date: | 2020-07-08 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structural basis for STAT2 suppression by flavivirus NS5.
Nat.Struct.Mol.Biol., 27, 2020
|
|
7T3D
 
 | | CryoEM map of anchor 222-1C06 Fab and lateral patch 2B05 Fab binding H1 HA | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 222-1C06 mAb heavy chain, ... | | Authors: | Han, J, Ward, A.B. | | Deposit date: | 2021-12-07 | | Release date: | 2022-01-12 | | Last modified: | 2022-03-30 | | Method: | ELECTRON MICROSCOPY (3.38 Å) | | Cite: | Broadly neutralizing antibodies target a haemagglutinin anchor epitope.
Nature, 602, 2022
|
|
8FRJ
 
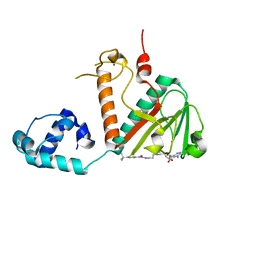 | | Structure of nsp14 N7-MethylTransferase domain fused with TELSAM bound to SGC0946 | | Descriptor: | 5-bromo-7-{5-[(3-{[(4-tert-butylphenyl)carbamoyl]amino}propyl)(propan-2-yl)amino]-5-deoxy-beta-D-ribofuranosyl}-7H-pyrrolo[2,3-d]pyrimidin-4-amine, Transcription factor ETV6,Guanine-N7 methyltransferase nsp14 chimera, ZINC ION | | Authors: | Kottur, J, Aggarwal, A.K. | | Deposit date: | 2023-01-07 | | Release date: | 2023-07-26 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Structures of SARS-CoV-2 N7-methyltransferase with DOT1L and PRMT7 inhibitors provide a platform for new antivirals.
Plos Pathog., 19, 2023
|
|
8FRK
 
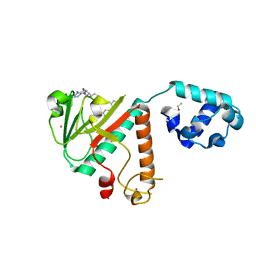 | |
6UX2
 
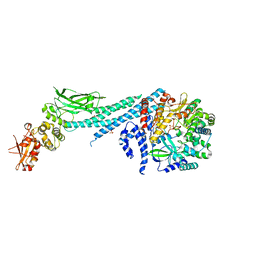 | | Crystal structure of ZIKV RdRp in complex with STAT2 | | Descriptor: | Nonstructural Protein 5, SULFATE ION, Signal transducer and activator of transcription 2, ... | | Authors: | Wang, B, Song, J. | | Deposit date: | 2019-11-06 | | Release date: | 2020-07-08 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.01 Å) | | Cite: | Structural basis for STAT2 suppression by flavivirus NS5.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6E7G
 
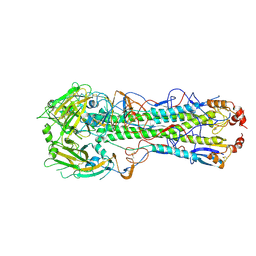 | |
6E7H
 
 | |
2LWE
 
 | |
2LWD
 
 | |
6N34
 
 | | Crystal structure of the BTB domain of Human NS1-BP | | Descriptor: | Influenza virus NS1A-binding protein | | Authors: | Zhang, K, Shang, G, Padavannil, A, Fontoura, B, Chook, Y.M. | | Deposit date: | 2018-11-14 | | Release date: | 2018-12-12 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural-functional interactions of NS1-BP protein with the splicing and mRNA export machineries for viral and host gene expression.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6N3H
 
 | | Crystal structure of Kelch domain of the human NS1 binding protein | | Descriptor: | Influenza virus NS1A-binding protein | | Authors: | Zhang, K, Shang, G, Padavannil, A, Fontoura, B.M.A, Chook, Y.M. | | Deposit date: | 2018-11-15 | | Release date: | 2018-12-12 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural-functional interactions of NS1-BP protein with the splicing and mRNA export machineries for viral and host gene expression.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6N5A
 
 | |
5BXZ
 
 | | H17 Bat Influenza NS1 RNA Binding Domain | | Descriptor: | Non-structural protein 1 | | Authors: | Kerry, P.S, Hale, B.G. | | Deposit date: | 2015-06-09 | | Release date: | 2015-08-19 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Novel Bat Influenza Virus NS1 Proteins Bind Double-Stranded RNA and Antagonize Host Innate Immunity.
J.Virol., 89, 2015
|
|
5BY1
 
 | | H18 Bat Influenza NS1 RNA Binding Domain | | Descriptor: | DI(HYDROXYETHYL)ETHER, Non-structural protein 1, PENTAETHYLENE GLYCOL | | Authors: | Kerry, P.S, Hale, B.G. | | Deposit date: | 2015-06-09 | | Release date: | 2015-08-19 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.751 Å) | | Cite: | Novel Bat Influenza Virus NS1 Proteins Bind Double-Stranded RNA and Antagonize Host Innate Immunity.
J.Virol., 89, 2015
|
|
8DIC
 
 | |
8DIF
 
 | |
8DIB
 
 | |
