1D5S
 
 | | CRYSTAL STRUCTURE OF CLEAVED ANTITRYPSIN POLYMER | | Descriptor: | P1-ARG ANTITRYPSIN | | Authors: | Dunstone, M.A, Dai, W, Whisstock, J.C, Rossjohn, J, Pike, R.N, Feil, S.C, Le Bonneic, B.F, Parker, M.W, Bottomley, S.P. | | Deposit date: | 1999-10-11 | | Release date: | 2000-04-02 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Cleaved antitrypsin polymers at atomic resolution.
Protein Sci., 9, 2000
|
|
6DLW
 
 | | Complement component polyC9 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Complement component C9, beta-D-mannopyranose | | Authors: | Dunstone, M.A, Spicer, B.A, Law, R.H.P. | | Deposit date: | 2018-06-03 | | Release date: | 2018-09-12 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | The first transmembrane region of complement component-9 acts as a brake on its self-assembly.
Nat Commun, 9, 2018
|
|
4OEJ
 
 | | Structure of membrane binding protein pleurotolysin B from Pleurotus ostreatus | | Descriptor: | ACETATE ION, CHLORIDE ION, GLYCEROL, ... | | Authors: | Dunstone, M.A, Caradoc-Davies, T.T, Whisstock, J.C, Law, R.H.P. | | Deposit date: | 2014-01-13 | | Release date: | 2015-02-18 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Conformational Changes during Pore Formation by the Perforin-Related Protein Pleurotolysin.
Plos Biol., 13, 2015
|
|
4OEB
 
 | | Structure of membrane binding protein pleurotolysin A from Pleurotus ostreatus | | Descriptor: | Pleurotolysin A, SULFATE ION | | Authors: | Dunstone, M.A, Caradoc-Davies, T.T, Whisstock, J.C, Law, R.H.P. | | Deposit date: | 2014-01-12 | | Release date: | 2015-02-18 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Conformational Changes during Pore Formation by the Perforin-Related Protein Pleurotolysin.
Plos Biol., 13, 2015
|
|
5FMW
 
 | | The poly-C9 component of the Complement Membrane Attack Complex | | Descriptor: | POLYC9 | | Authors: | Dudkina, N.V, Spicer, B.A, Reboul, C.F, Conroy, P.J, Lukoyanova, N, Elmlund, H, Law, R.H.P, Ekkel, S.M, Kondos, S.C, Goode, R.J.A, Ramm, G, Whisstock, J.C, Saibil, H.R, Dunstone, M.A. | | Deposit date: | 2015-11-10 | | Release date: | 2016-02-17 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (6.7 Å) | | Cite: | Structure of the Poly-C9 Component of the Complement Membrane Attack Complex
Nat.Commun., 7, 2016
|
|
4V3N
 
 | | Membrane bound pleurotolysin prepore (TMH2 strand lock) trapped with engineered disulphide cross-link | | Descriptor: | PLEUROTOLYSIN A, PLEUROTOLYSIN B | | Authors: | Lukoyanova, N, Kondos, S.C, Farabella, I, Law, R.H.P, Reboul, C.F, Caradoc-Davies, T.T, Spicer, B.A, Kleifeld, O, Perugini, M, Ekkel, S, Hatfaludi, T, Oliver, K, Hotze, E.M, Tweten, R.K, Whisstock, J.C, Topf, M, Dunstone, M.A, Saibil, H.R. | | Deposit date: | 2014-10-20 | | Release date: | 2015-02-18 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (14 Å) | | Cite: | Conformational Changes During Pore Formation by the Perforin-Related Protein Pleurotolysin.
Plos Biol., 13, 2015
|
|
4V3A
 
 | | Membrane bound pleurotolysin prepore (TMH1 lock) trapped with engineered disulphide cross-link | | Descriptor: | PLEUROTOLYSIN A, PLEUROTOLYSIN B | | Authors: | Lukoyanova, N, Kondos, S.C, Farabella, I, Law, R.H.P, Reboul, C.F, CaradocDavies, T.T, Spicer, B.A, Kleifeld, O, Perugini, M, Ekkel, S, Hatfaludi, T, Oliver, K, Hotze, E.M, Tweten, R.K, Whisstock, J.C, Topf, M, Dunstone, M.A, Saibil, H.R. | | Deposit date: | 2014-10-17 | | Release date: | 2015-02-18 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (15 Å) | | Cite: | Conformational Changes During Pore Formation by the Perforin-Related Protein Pleurotolysin.
Plos Biol., 13, 2015
|
|
4V3M
 
 | | Membrane bound pleurotolysin prepore (TMH2 helix lock) trapped with engineered disulphide cross-link | | Descriptor: | PLEUROTOLYSIN A, PLEUROTOLYSIN B | | Authors: | Lukoyanova, N, Kondos, S.C, Farabella, I, Law, R.H.P, Reboul, C.F, Caradoc-Davies, T.T, Spicer, B.A, Kleifeld, O, Perugini, M, Ekkel, S, Hatfaludi, T, Oliver, K, Hotze, E.M, Tweten, R.K, Whisstock, J.C, Topf, M, Dunstone, M.A, Saibil, H.R. | | Deposit date: | 2014-10-20 | | Release date: | 2015-02-18 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (17 Å) | | Cite: | Conformational Changes During Pore Formation by the Perforin-Related Protein Pleurotolysin.
Plos Biol., 13, 2015
|
|
4V2T
 
 | | Membrane embedded pleurotolysin pore with 13 fold symmetry | | Descriptor: | PLEUROTOLYSIN A, PLEUROTOLYSIN B | | Authors: | Lukoyanova, N, Kondos, S.C, Farabella, I, Law, R.H.P, Reboul, C.F, Caradoc-Davies, T.T, Spicer, B.A, Kleifeld, O, Perugini, M, Ekkel, S, Hatfaludi, T, Oliver, K, Hotze, E.M, Tweten, R.K, Whisstock, J.C, Topf, M, Dunstone, M.A, Saibil, H.R. | | Deposit date: | 2014-10-15 | | Release date: | 2015-02-18 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (11 Å) | | Cite: | Conformational Changes During Pore Formation by the Perforin-Related Protein Pleurotolysin.
Plos Biol., 13, 2015
|
|
2QP2
 
 | | Structure of a MACPF/perforin-like protein | | Descriptor: | CALCIUM ION, Unknown protein | | Authors: | Rosado, C.J, Buckle, A.M, Law, R.H.P, Butcher, R.E, Kan, W.T, Bird, C.H, Ung, K, Browne, K.A, Baran, K, Bashtannyk-Puhalovich, T.A, Faux, N.G, Wong, W, Porter, C.J, Pike, R.N, Ellisdon, A.M, Pearce, M.C, Bottomley, S.P, Emsley, J, Smith, A.I, Rossjohn, J, Hartland, E.L, Voskoboinik, I, Trapani, J.A, Bird, P.I, Dunstone, M.A, Whisstock, J.C. | | Deposit date: | 2007-07-22 | | Release date: | 2007-09-04 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A common fold mediates vertebrate defense and bacterial attack
Science, 317, 2007
|
|
3L3H
 
 | | X-ray crystal structure of the F6A mutant of influenza A acid polymerase epitope PA224 bound to murine H2-Db MHC | | Descriptor: | 10-mer peptide from Polymerase acidic protein, Beta-2-microglobulin, H-2 class I histocompatibility antigen, ... | | Authors: | Welland, A, Clements, C.S, Dunstone, M.A, Rossjohn, J. | | Deposit date: | 2009-12-17 | | Release date: | 2010-03-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Constraints within major histocompatibility complex class I restricted peptides: presentation and consequences for T-cell recognition
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
1YN7
 
 | | Crystal structure of a mouse MHC class I protein, H2-Db, in complex with a mutated peptide (R7A) of the influenza A acid polymerase | | Descriptor: | 10-mer peptide from RNA-directed RNA polymerase subunit P2, Beta-2-microglobulin, H-2 class I histocompatibility antigen, ... | | Authors: | Turner, S.J, Kedzierska, K, Komodromou, H, La Gruta, N.L, Dunstone, M.A, Webb, A.I, Webby, R, Walden, H, Xie, W, McCluskey, J, Purcell, A.W, Rossjohn, J, Doherty, P.C. | | Deposit date: | 2005-01-24 | | Release date: | 2005-06-28 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Lack of prominent peptide-major histocompatibility complex features limits repertoire diversity in virus-specific CD8+ T cell populations
Nat.Immunol., 6, 2005
|
|
1YN6
 
 | | Crystal structure of a mouse MHC class I protein, H2-Db, in complex with a peptide from the influenza A acid polymerase | | Descriptor: | 10-mer peptide from RNA-directed RNA polymerase subunit P2, Beta-2-microglobulin, H-2 class I histocompatibility antigen, ... | | Authors: | Turner, S.J, Kedzierska, K, Komodromou, H, La Gruta, N.L, Dunstone, M.A, Webb, A.I, Webby, R, Walden, H, Xie, W, McCluskey, J, Purcell, A.W, Rossjohn, J, Doherty, P.C. | | Deposit date: | 2005-01-23 | | Release date: | 2005-06-28 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Lack of prominent peptide-major histocompatibility complex features limits repertoire diversity in virus-specific CD8+ T cell populations
Nat.Immunol., 6, 2005
|
|
1YDP
 
 | | 1.9A crystal structure of HLA-G | | Descriptor: | Beta-2-microglobulin, CHLORIDE ION, COBALT (II) ION, ... | | Authors: | Clements, C.S, Kjer-nielsen, L, Kostenko, L, Hoare, H.L, Dunstone, M.A, Moses, E, Freed, K, Brooks, A.G, Rossjohn, J, Mccluskey, J. | | Deposit date: | 2004-12-25 | | Release date: | 2005-03-08 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of HLA-G: A nonclassical MHC class I molecule expressed at the fetal-maternal interface
PROC.NATL.ACAD.SCI.USA, 102, 2005
|
|
2ZSW
 
 | | Crystal structure of H-2Kb in complex with the Q600Y variant of JHMV epitope S598 | | Descriptor: | 8-mer peptide from spike glycoprotein, Beta-2-microglobulin, H-2 class I histocompatibility antigen, ... | | Authors: | Theodossis, A, Dunstone, M.A, Rossjohn, J. | | Deposit date: | 2008-09-18 | | Release date: | 2008-11-04 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Prevention of cytotoxic T cell escape using a heteroclitic subdominant viral T cell determinant.
Plos Pathog., 4, 2008
|
|
2ZOL
 
 | | Crystal structure of H-2Db in complex with the W513S variant of JHMV epitope S510 | | Descriptor: | 9-meric peptide from Spike glycoprotein, Beta-2-microglobulin, H-2 class I histocompatibility antigen, ... | | Authors: | Theodossis, A, Dunstone, M.A, Rossjohn, J. | | Deposit date: | 2008-05-22 | | Release date: | 2008-06-10 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural and biological basis of CTL escape in coronavirus-infected mice.
J Immunol., 180, 2008
|
|
2ZOK
 
 | | Crystal structure of H-2Db in complex with JHMV epitope S510 | | Descriptor: | 9-meric peptide from Spike glycoprotein, Beta-2-microglobulin, GLYCEROL, ... | | Authors: | Theodossis, A, Dunstone, M.A, Rossjohn, J. | | Deposit date: | 2008-05-22 | | Release date: | 2008-06-10 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural and biological basis of CTL escape in coronavirus-infected mice.
J Immunol., 180, 2008
|
|
2ZSV
 
 | | Crystal structure of H-2Kb in complex with JHMV epitope S598 | | Descriptor: | 8-mer peptide from spike glycoprotein, Beta-2-microglobulin, CYSTEINE, ... | | Authors: | Theodossis, A, Dunstone, M.A, Rossjohn, J. | | Deposit date: | 2008-09-18 | | Release date: | 2008-11-04 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Prevention of cytotoxic T cell escape using a heteroclitic subdominant viral T cell determinant.
Plos Pathog., 4, 2008
|
|
3BUY
 
 | | MHC-I in complex with peptide | | Descriptor: | Beta-2-microglobulin, H-2 class I histocompatibility antigen, D-B alpha chain, ... | | Authors: | Rossjohn, J, La Gruta, N.L, Purcell, A.W, Turner, S.J, Dunstone, M.A. | | Deposit date: | 2008-01-03 | | Release date: | 2008-03-25 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Epitope-specific TCRbeta repertoire diversity imparts no functional advantage on the CD8+ T cell response to cognate viral peptides
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
4OV8
 
 | | Crystal Structure of the TMH1-lock mutant of the mature form of pleurotolysin B | | Descriptor: | CHLORIDE ION, GLYCEROL, Pleurotolysin B, ... | | Authors: | Kondos, S.C, Law, R.H.P, Whisstock, J.C, Dunstone, M.A. | | Deposit date: | 2014-02-20 | | Release date: | 2015-02-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Conformational Changes during Pore Formation by the Perforin-Related Protein Pleurotolysin
Plos Biol., 13, 2015
|
|
1S9Y
 
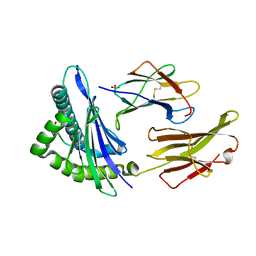 | | Crystal Structure Analysis of NY-ESO-1 epitope analogue, SLLMWITQS, in complex with HLA-A2 | | Descriptor: | Beta-2-microglobulin, HLA class I histocompatibility antigen, A-2 alpha chain, ... | | Authors: | Webb, A.I, Dunstone, M.A, Chen, W, Aguilar, M.I, Chen, Q, Chang, L, Kjer-Nielsen, L, Beddoe, T, McCluskey, J, Rossjohn, J, Purcell, A.W. | | Deposit date: | 2004-02-05 | | Release date: | 2004-09-28 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Functional and structural characteristics of NY-ESO-1-related HLA A2-restricted epitopes and the design of a novel immunogenic analogue
J.Biol.Chem., 279, 2004
|
|
1SY6
 
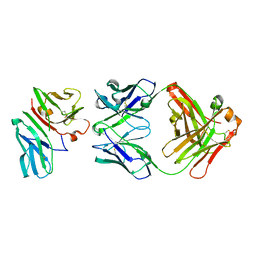 | | Crystal Structure of CD3gammaepsilon Heterodimer in Complex with OKT3 Fab Fragment | | Descriptor: | OKT3 Fab heavy chain, OKT3 Fab light chain, T-cell surface glycoprotein CD3 gamma/epsilon chain | | Authors: | Kjer-Nielsen, L, Dunstone, M.A, Kostenko, L, Ely, L.K, Beddoe, T, Misfud, N.A, Purcell, A.W, Brooks, A.G, McCluskey, J, Rossjohn, J. | | Deposit date: | 2004-03-31 | | Release date: | 2004-05-25 | | Last modified: | 2018-04-04 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of the human T cell receptor CD3(epsilon)(gamma) heterodimer complexed to the therapeutic mAb OKT3.
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
1S9X
 
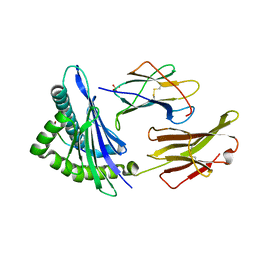 | | Crystal Structure Analysis of NY-ESO-1 epitope analogue, SLLMWITQA, in complex with HLA-A2 | | Descriptor: | Beta-2-microglobulin, HLA class I histocompatibility antigen, A-2 alpha chain, ... | | Authors: | Webb, A.I, Dunstone, M.A, Chen, W, Aguilar, M.I, Chen, Q, Chang, L, Kjer-Nielsen, L, Beddoe, T, McCluskey, J, Rossjohn, J, Purcell, A.W. | | Deposit date: | 2004-02-05 | | Release date: | 2004-09-28 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Functional and structural characteristics of NY-ESO-1-related HLA A2-restricted epitopes and the design of a novel immunogenic analogue
J.Biol.Chem., 279, 2004
|
|
1S9W
 
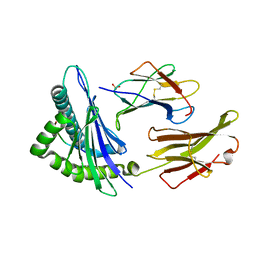 | | Crystal Structure Analysis of NY-ESO-1 epitope, SLLMWITQC, in complex with HLA-A2 | | Descriptor: | Beta-2-microglobulin, HLA class I histocompatibility antigen, A-2 alpha chain, ... | | Authors: | Webb, A.I, Dunstone, M.A, Chen, W, Aguilar, M.I, Chen, Q, Chang, L, Kjer-Nielsen, L, Beddoe, T, McCluskey, J, Rossjohn, J, Purcell, A.W. | | Deposit date: | 2004-02-05 | | Release date: | 2004-09-28 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Functional and structural characteristics of NY-ESO-1-related HLA A2-restricted epitopes and the design of a novel immunogenic analogue
J.Biol.Chem., 279, 2004
|
|
1T0M
 
 | | Conformational switch in polymorphic H-2K molecules containing an HSV peptide | | Descriptor: | Beta-2-microglobulin, Glycoprotein B, H-2 class I histocompatibility antigen, ... | | Authors: | Webb, A.I, Borg, N.A, Dunstone, M.A, Kjer-Nielsen, L, Beddoe, T, McCluskey, J, Carbone, F.R, Bottomley, S.P, Purcell, A.W, Rossjohn, J. | | Deposit date: | 2004-04-12 | | Release date: | 2004-11-23 | | Last modified: | 2019-11-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The structure of H-2K(b) and K(bm8) complexed to a herpes simplex virus determinant: evidence for a conformational switch that governs T cell repertoire selection and viral resistance.
J Immunol., 173, 2004
|
|
