5FXH
 
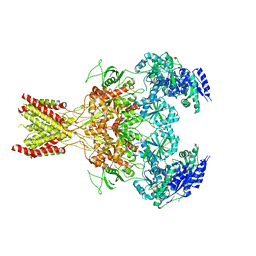 | | GluN1b-GluN2B NMDA receptor in non-active-1 conformation | | Descriptor: | N-METHYL-D-ASPARTATE RECEPTOR GLUN1, N-METHYL-D-ASPARTATE RECEPTOR GLUN2B | | Authors: | Tajima, N, Karakas, E, Grant, T, Simorowski, N, Diaz-Avalos, R, Grigorieff, N, Furukawa, H. | | Deposit date: | 2016-03-02 | | Release date: | 2016-05-11 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (5 Å) | | Cite: | Activation of Nmda Receptors and the Mechanism of Inhibition by Ifenprodil.
Nature, 534, 2016
|
|
5B3J
 
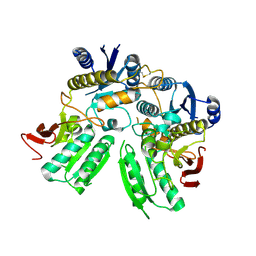 | | Activation of NMDA receptors and the mechanism of inhibition by ifenprodil | | Descriptor: | Fab, heavy chain, light chain, ... | | Authors: | Tajima, N, Karakas, E, Grant, T, Simorowski, N, Diaz-Avalos, R, Grigorieff, N, Furukawa, H. | | Deposit date: | 2016-03-01 | | Release date: | 2016-05-11 | | Last modified: | 2022-03-23 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Activation of NMDA receptors and the mechanism of inhibition by ifenprodil
Nature, 534, 2016
|
|
5U9F
 
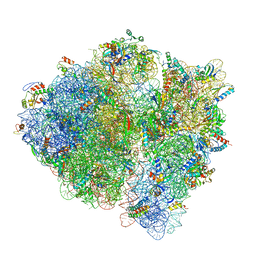 | | 3.2 A cryo-EM ArfA-RF2 ribosome rescue complex (Structure II) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Demo, G, Svidritskiy, E, Madireddy, R, Diaz-Avalos, R, Grant, T, Grigorieff, N, Sousa, D, Korostelev, A.A. | | Deposit date: | 2016-12-16 | | Release date: | 2017-03-22 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Mechanism of ribosome rescue by ArfA and RF2.
Elife, 6, 2017
|
|
5U9G
 
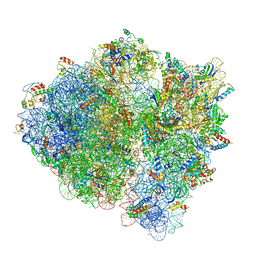 | | 3.2 A cryo-EM ArfA-RF2 ribosome rescue complex (Structure I) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Demo, G, Svidritskiy, E, Madireddy, R, Diaz-Avalos, R, Grant, T, Grigorieff, N, Sousa, D, Korostelev, A.A. | | Deposit date: | 2016-12-16 | | Release date: | 2017-03-22 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Mechanism of ribosome rescue by ArfA and RF2.
Elife, 6, 2017
|
|
3J2U
 
 | | Kinesin-13 KLP10A HD in complex with CS-tubulin and a microtubule | | Descriptor: | Kinesin-like protein Klp10A, Tubulin alpha-1A chain, Tubulin beta-2B chain | | Authors: | Asenjo, A.B, Chatterjee, C, Tan, D, DePaoli, V, Rice, W.J, Diaz-Avalos, R, Silvestry, M, Sosa, H. | | Deposit date: | 2013-01-10 | | Release date: | 2013-03-06 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (10.8 Å) | | Cite: | Structural model for tubulin recognition and deformation by Kinesin-13 microtubule depolymerases.
Cell Rep, 3, 2013
|
|
6AWB
 
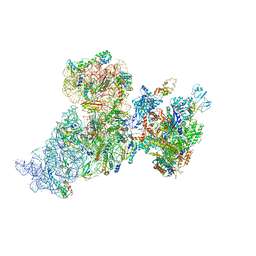 | | Structure of 30S ribosomal subunit and RNA polymerase complex in non-rotated state | | Descriptor: | 16S rRNA, 30S ribosomal protein S1, 30S ribosomal protein S10, ... | | Authors: | Demo, G, Rasouly, A, Vasilyev, N, Loveland, A.B, Diaz-Avalos, R, Grigorieff, N, Nudler, E, Korostelev, A.A. | | Deposit date: | 2017-09-05 | | Release date: | 2017-10-18 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (6.7 Å) | | Cite: | Structure of RNA polymerase bound to ribosomal 30S subunit.
Elife, 6, 2017
|
|
6AWD
 
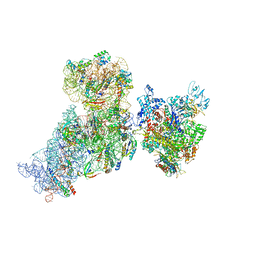 | | Structure of 30S (S1 depleted) ribosomal subunit and RNA polymerase complex | | Descriptor: | 16S rRNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | Authors: | Demo, G, Rasouly, A, Vasilyev, N, Loveland, A.B, Diaz-Avalos, R, Grigorieff, N, Nudler, E, Korostelev, A.A. | | Deposit date: | 2017-09-05 | | Release date: | 2017-10-18 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (8.1 Å) | | Cite: | Structure of RNA polymerase bound to ribosomal 30S subunit.
Elife, 6, 2017
|
|
6AWC
 
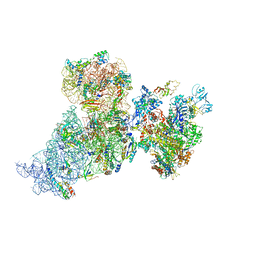 | | Structure of 30S ribosomal subunit and RNA polymerase complex in rotated state | | Descriptor: | 16S rRNA, 30S ribosomal protein S1, 30S ribosomal protein S10, ... | | Authors: | Demo, G, Rasouly, A, Vasilyev, N, Loveland, A.B, Diaz-Avalos, R, Grigorieff, N, Nudler, E, Korostelev, A.A. | | Deposit date: | 2017-09-05 | | Release date: | 2017-10-18 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (7.9 Å) | | Cite: | Structure of RNA polymerase bound to ribosomal 30S subunit.
Elife, 6, 2017
|
|
5FXJ
 
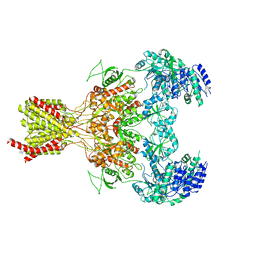 | | GluN1b-GluN2B NMDA receptor structure-Class X | | Descriptor: | GLUTAMATE RECEPTOR IONOTROPIC, NMDA 1, NMDA 2B | | Authors: | Tajima, N, Karakas, E, Grant, T, Simorowski, N, Diaz-Avalos, R, Grigorieff, N, Furukawa H, H. | | Deposit date: | 2016-03-02 | | Release date: | 2016-05-25 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (6.25 Å) | | Cite: | Activation of Nmda Receptors and the Mechanism of Inhibition by Ifenprodil.
Nature, 534, 2016
|
|
5FXI
 
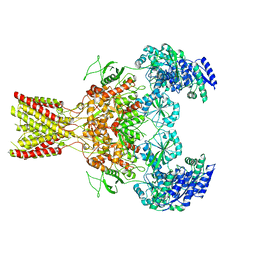 | | GluN1b-GluN2B NMDA receptor structure in non-active-2 conformation | | Descriptor: | N-METHYL-D-ASPARTATE RECEPTOR GLUN1, N-METHYL-D-ASPARTATE RECEPTOR GLUN2B | | Authors: | Tajima, N, Karakas, E, Grant, T, Simorowski, N, Diaz-Avalos, R, Grigorieff, N, Furukawa, H. | | Deposit date: | 2016-03-02 | | Release date: | 2016-05-11 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (6.4 Å) | | Cite: | Activation of Nmda Receptors and the Mechanism of Inhibition by Ifenprodil.
Nature, 534, 2016
|
|
5FXK
 
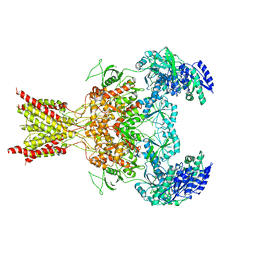 | | GluN1b-GluN2B NMDA receptor structure-Class Y | | Descriptor: | N-METHYL-D-ASPARTATE RECEPTOR GLUN1, N-METHYL-D-ASPARTATE RECEPTOR GLUN2B | | Authors: | Tajima, N, Karakas, E, Grant, T, Simorowski, N, Diaz-Avalos, R, Grigorieff, N, Furukawa, H. | | Deposit date: | 2016-03-02 | | Release date: | 2016-05-11 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (6.4 Å) | | Cite: | Activation of Nmda Receptors and the Mechanism of Inhibition by Ifenprodil.
Nature, 534, 2016
|
|
5FXG
 
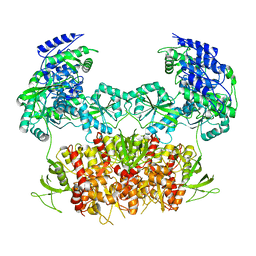 | | GLUN1B-GLUN2B NMDA RECEPTOR IN ACTIVE CONFORMATION | | Descriptor: | N-METHYL-D-ASPARTATE RECEPTOR GLUN1, N-METHYL-D-ASPARTATE RECEPTOR GLUN2B | | Authors: | Tajima, N, Karakas, E, Grant, T, Simorowski, N, Diaz-Avalos, R, Grigorieff, N, Furukawa, H. | | Deposit date: | 2016-03-02 | | Release date: | 2016-05-11 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (6.8 Å) | | Cite: | Activation of Nmda Receptors and the Mechanism of Inhibition by Ifenprodil.
Nature, 534, 2016
|
|
4GOY
 
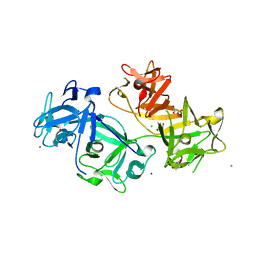 | | The crystal structure of human fascin 1 K41A mutant | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, BROMIDE ION, CHLORIDE ION, ... | | Authors: | Yang, S.Y, Huang, F.K, Huang, J, Chen, S, Jakoncic, J, Leo-Macias, A, Diaz-Avalos, R, Chen, L, Zhang, J.J, Huang, X.Y. | | Deposit date: | 2012-08-20 | | Release date: | 2012-11-28 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Molecular mechanism of fascin function in filopodial formation.
J.Biol.Chem., 288, 2013
|
|
4GP3
 
 | | The crystal structure of human fascin 1 K358A mutant | | Descriptor: | BROMIDE ION, CHLORIDE ION, Fascin, ... | | Authors: | Yang, S.Y, Huang, F.K, Huang, J, Chen, S, Jakoncic, J, Leo-Macias, A, Diaz-Avalos, R, Chen, L, Zhang, J.J, Huang, X.Y. | | Deposit date: | 2012-08-20 | | Release date: | 2012-11-28 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Molecular mechanism of fascin function in filopodial formation.
J.Biol.Chem., 288, 2013
|
|
4GP0
 
 | | The crystal structure of human fascin 1 R149A K150A R151A mutant | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, BROMIDE ION, ... | | Authors: | Yang, S.Y, Huang, F.K, Huang, J, Chen, S, Jakoncic, J, Leo-Macias, A, Diaz-Avalos, R, Chen, L, Zhang, J.J, Huang, X.Y. | | Deposit date: | 2012-08-20 | | Release date: | 2012-11-28 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Molecular mechanism of fascin function in filopodial formation.
J.Biol.Chem., 288, 2013
|
|
4GOV
 
 | | The crystal structure of human fascin 1 S39D mutant | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, BROMIDE ION, CHLORIDE ION, ... | | Authors: | Yang, S.Y, Huang, F.K, Huang, J, Chen, S, Jakoncic, J, Leo-Macias, A, Diaz-Avalos, R, Chen, L, Zhang, J.J, Huang, X.Y. | | Deposit date: | 2012-08-20 | | Release date: | 2012-11-28 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Molecular mechanism of fascin function in filopodial formation.
J.Biol.Chem., 288, 2013
|
|
6U1X
 
 | | Structure of the Vesicular Stomatitis Virus L Protein in Complex with Its Phosphoprotein Cofactor (3.0 A resolution) | | Descriptor: | Phosphoprotein, RNA-directed RNA polymerase L, ZINC ION | | Authors: | Jenni, S, Bloyet, L.M, Dias-Avalos, R, Liang, B, Wheelman, S.P.J, Grigorieff, N, Harrison, S.C. | | Deposit date: | 2019-08-17 | | Release date: | 2020-01-22 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structure of the Vesicular Stomatitis Virus L Protein in Complex with Its Phosphoprotein Cofactor.
Cell Rep, 30, 2020
|
|
3RIU
 
 | |
