6QSZ
 
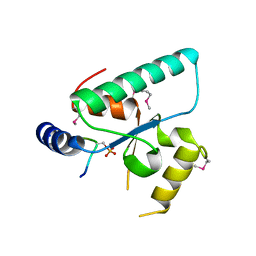 | | Crystal structure of the Sir4 H-BRCT domain in complex with Esc1 pS1450 peptide | | Descriptor: | CHLORIDE ION, Regulatory protein SIR4, Silent chromatin protein ESC1 | | Authors: | Deshpande, I, Keusch, J.J, Challa, K, Iesmantavicius, V, Gasser, S.M, Gut, H. | | Deposit date: | 2019-02-22 | | Release date: | 2019-09-18 | | Last modified: | 2019-10-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The Sir4 H-BRCT domain interacts with phospho-proteins to sequester and repress yeast heterochromatin.
Embo J., 38, 2019
|
|
6RR0
 
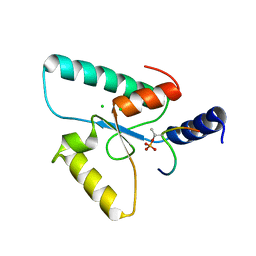 | | Crystal structure of the Sir4 H-BRCT domain in complex with Ubp10 pT123 peptide | | Descriptor: | CHLORIDE ION, Regulatory protein SIR4, Ubiquitin carboxyl-terminal hydrolase 10 | | Authors: | Deshpande, I, Keusch, J.J, Challa, K, Iesmantavicius, V, Gasser, S.M, Gut, H. | | Deposit date: | 2019-05-16 | | Release date: | 2019-09-18 | | Last modified: | 2019-10-23 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | The Sir4 H-BRCT domain interacts with phospho-proteins to sequester and repress yeast heterochromatin.
Embo J., 38, 2019
|
|
6RRV
 
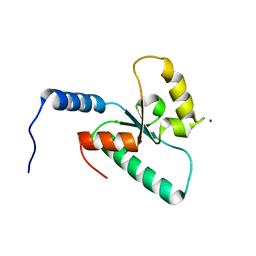 | | Crystal structure of the Sir4 H-BRCT domain | | Descriptor: | BROMIDE ION, CHLORIDE ION, Regulatory protein SIR4 | | Authors: | Deshpande, I, Keusch, J.J, Challa, K, Iesmantavicius, V, Gasser, S.M, Gut, H. | | Deposit date: | 2019-05-20 | | Release date: | 2019-09-18 | | Last modified: | 2019-10-23 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | The Sir4 H-BRCT domain interacts with phospho-proteins to sequester and repress yeast heterochromatin.
Embo J., 38, 2019
|
|
5OMB
 
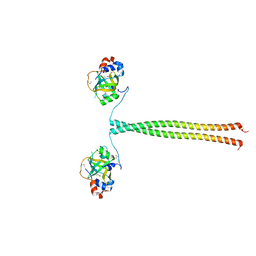 | | Crystal structure of K. lactis Ddc2 N-terminus in complex with S. cerevisiae Rfa1 N-OB domain | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DNA damage checkpoint protein LCD1, ... | | Authors: | Deshpande, I, Seeber, A, Shimada, K, Keusch, J.J, Gut, H, Gasser, S.M. | | Deposit date: | 2017-07-28 | | Release date: | 2017-10-25 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Structural Basis of Mec1-Ddc2-RPA Assembly and Activation on Single-Stranded DNA at Sites of Damage.
Mol. Cell, 68, 2017
|
|
5OMD
 
 | | Crystal structure of S. cerevisiae Ddc2 N-terminal coiled-coil domain | | Descriptor: | DNA damage checkpoint protein LCD1 | | Authors: | Deshpande, I, Seeber, A, Shimada, K, Keusch, J.J, Gut, H, Gasser, S.M. | | Deposit date: | 2017-07-28 | | Release date: | 2017-10-25 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Basis of Mec1-Ddc2-RPA Assembly and Activation on Single-Stranded DNA at Sites of Damage.
Mol. Cell, 68, 2017
|
|
5OMC
 
 | | Crystal structure of K. lactis Ddc2 N-terminus in complex with S. cerevisiae Rfa1 (K45E mutant) N-OB domain | | Descriptor: | CHLORIDE ION, DNA damage checkpoint protein LCD1, Replication factor A protein 1 | | Authors: | Deshpande, I, Seeber, A, Shimada, K, Keusch, J.J, Gut, H, Gasser, S.M. | | Deposit date: | 2017-07-28 | | Release date: | 2017-10-25 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Structural Basis of Mec1-Ddc2-RPA Assembly and Activation on Single-Stranded DNA at Sites of Damage.
Mol. Cell, 68, 2017
|
|
6QTM
 
 | | Crystal structure of the Sir4 H-BRCT domain in complex with Ty5 pS1095 peptide | | Descriptor: | Regulatory protein SIR4, Ribonuclease H, SULFATE ION | | Authors: | Gut, H, Deshpande, I, Keusch, J.J, Challa, K, Iesmantavicius, V, Gasser, S.M. | | Deposit date: | 2019-02-25 | | Release date: | 2019-09-18 | | Last modified: | 2021-08-11 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The Sir4 H-BRCT domain interacts with phospho-proteins to sequester and repress yeast heterochromatin.
Embo J., 38, 2019
|
|
5M1X
 
 | | Crystal structure of S. cerevisiae Rfa1 N-OB domain mutant (K45E) | | Descriptor: | Replication factor A protein 1 | | Authors: | Seeber, A, Hegnauer, A.M, Hustedt, N, Deshpande, I, Poli, J, Eglinger, J, Pasero, P, Gut, H, Shinohara, M, Hopfner, K.P, Shimada, K, Gasser, S.M. | | Deposit date: | 2016-10-11 | | Release date: | 2016-12-07 | | Last modified: | 2016-12-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | RPA Mediates Recruitment of MRX to Forks and Double-Strand Breaks to Hold Sister Chromatids Together.
Mol. Cell, 64, 2016
|
|
6O3C
 
 | | Crystal structure of active Smoothened bound to SAG21k, cholesterol, and NbSmo8 | | Descriptor: | (2S)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 2-acetamido-2-deoxy-beta-D-glucopyranose, 3-chloro-4,7-difluoro-N-{[2-methoxy-5-(pyridin-4-yl)phenyl]methyl}-N-[trans-4-(methylamino)cyclohexyl]-1-benzothiophene-2-carboxamide, ... | | Authors: | Deshpande, I.S, Liang, J, Hedeen, D, Roberts, K.J, Zhang, Y, Ha, B, Latorraca, N.R, Faust, B, Dror, R.O, Beachy, P.A, Myers, B.R, Manglik, A. | | Deposit date: | 2019-02-26 | | Release date: | 2019-07-03 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Smoothened stimulation by membrane sterols drives Hedgehog pathway activity.
Nature, 571, 2019
|
|
8SMV
 
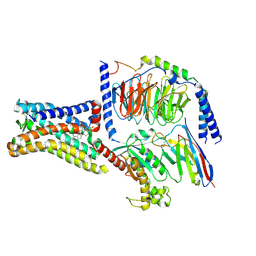 | | GPR161 Gs heterotrimer | | Descriptor: | CHOLESTEROL, G-protein coupled receptor 161, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Hoppe, N, Manglik, A, Harrison, S. | | Deposit date: | 2023-04-26 | | Release date: | 2024-02-21 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (2.74 Å) | | Cite: | GPR161 structure uncovers the redundant role of sterol-regulated ciliary cAMP signaling in the Hedgehog pathway.
Biorxiv, 2023
|
|
7KDT
 
 | |
7KQP
 
 | | Crystal structure of SARS-CoV-2 NSP3 macrodomain in complex with ADP-ribose (P43 crystal form) | | Descriptor: | Non-structural protein 3, [(2R,3S,4R,5R)-5-(6-AMINOPURIN-9-YL)-3,4-DIHYDROXY-OXOLAN-2-YL]METHYL [HYDROXY-[[(2R,3S,4R,5S)-3,4,5-TRIHYDROXYOXOLAN-2-YL]METHOXY]PHOSPHORYL] HYDROGEN PHOSPHATE | | Authors: | Correy, G.J, Young, I.D, Thompson, M.C, Fraser, J.S. | | Deposit date: | 2020-11-17 | | Release date: | 2020-12-09 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (0.88 Å) | | Cite: | Fragment binding to the Nsp3 macrodomain of SARS-CoV-2 identified through crystallographic screening and computational docking.
Sci Adv, 7, 2021
|
|
7KQW
 
 | | Crystal structure of SARS-CoV-2 NSP3 macrodomain (C2 crystal form, methylated) | | Descriptor: | Non-structural protein 3 | | Authors: | Correy, G.J, Young, I.D, Thompson, M.C, Fraser, J.S. | | Deposit date: | 2020-11-17 | | Release date: | 2020-12-09 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (0.93 Å) | | Cite: | Fragment binding to the Nsp3 macrodomain of SARS-CoV-2 identified through crystallographic screening and computational docking.
Sci Adv, 7, 2021
|
|
7KQO
 
 | |
7KR0
 
 | | Crystal structure of SARS-CoV-2 NSP3 macrodomain (C2 crystal form, 100 K) | | Descriptor: | Non-structural protein 3 | | Authors: | Correy, G.J, Young, I.D, Thompson, M.C, Fraser, J.S. | | Deposit date: | 2020-11-18 | | Release date: | 2020-12-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (0.77 Å) | | Cite: | Fragment binding to the Nsp3 macrodomain of SARS-CoV-2 identified through crystallographic screening and computational docking.
Sci Adv, 7, 2021
|
|
7KR1
 
 | | Crystal structure of SARS-CoV-2 NSP3 macrodomain (C2 crystal form, 310 K) | | Descriptor: | Non-structural protein 3 | | Authors: | Correy, G.J, Young, I.D, Thompson, M.C, Fraser, J.S. | | Deposit date: | 2020-11-18 | | Release date: | 2020-12-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Fragment binding to the Nsp3 macrodomain of SARS-CoV-2 identified through crystallographic screening and computational docking.
Sci Adv, 7, 2021
|
|
7KKJ
 
 | | Structure of anti-SARS-CoV-2 Spike nanobody mNb6 | | Descriptor: | CHLORIDE ION, SULFATE ION, Synthetic nanobody mNb6 | | Authors: | Schoof, M.S, Faust, B.F, Saunders, R.A, Sangwan, S, Rezelj, V, Hoppe, N, Boone, M, Billesboelle, C.B, Puchades, C, Azumaya, C.M, Kratochvil, H.T, Zimanyi, M, Desphande, I, Liang, J, Dickinson, S, Nguyen, H.C, Chio, C.M, Merz, G.E, Thompson, M.C, Diwanji, D, Schaefer, K, Anand, A.A, Dobzinski, N, Zha, B.S, Simoneau, C.R, Leon, K, White, K.M, Chio, U.S, Gupta, M, Jin, M, Li, F, Liu, Y, Zhang, K, Bulkley, D, Sun, M, Smith, A.M, Rizo, A.N, Moss, F, Brilot, A.F, Pourmal, S, Trenker, R, Pospiech, T, Gupta, S, Barsi-Rhyne, B, Belyy, V, Barile-Hill, A.W, Nock, S, Liu, Y, Krogan, N.J, Ralston, C.Y, Swaney, D.L, Garcia-Sastre, A, Ott, M, Vignuzzi, M, Walter, P, Manglik, A, QCRG Structural Biology Consortium | | Deposit date: | 2020-10-27 | | Release date: | 2020-11-25 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | An ultrapotent synthetic nanobody neutralizes SARS-CoV-2 by stabilizing inactive Spike.
Science, 370, 2020
|
|
7KKK
 
 | | SARS-CoV-2 Spike in complex with neutralizing nanobody Nb6 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Schoof, M.S, Faust, B.F, Saunders, R.A, Sangwan, S, Rezelj, V, Hoppe, N, Boone, M, Billesboelle, C.B, Puchades, C, Azumaya, C.M, Kratochvil, H.T, Zimanyi, M, Desphande, I, Liang, J, Dickinson, S, Nguyen, H.C, Chio, C.M, Merz, G.E, Thompson, M.C, Diwanji, D, Schaefer, K, Anand, A.A, Dobzinski, N, Zha, B.S, Simoneau, C.R, Leon, K, White, K.M, Chio, U.S, Gupta, M, Jin, M, Li, F, Liu, Y, Zhang, K, Bulkley, D, Sun, M, Smith, A.M, Rizo, A.N, Moss, F, Brilot, A.F, Pourmal, S, Trenker, R, Pospiech, T, Gupta, S, Barsi-Rhyne, B, Belyy, V, Barile-Hill, A.W, Nock, S, Liu, Y, Krogan, N.J, Ralston, C.Y, Swaney, D.L, Garcia-Sastre, A, Ott, M, Vignuzzi, M, Walter, P, Manglik, A, QCRG Structural Biology Consortium | | Deposit date: | 2020-10-27 | | Release date: | 2020-11-11 | | Last modified: | 2021-04-21 | | Method: | ELECTRON MICROSCOPY (3.03 Å) | | Cite: | An ultrapotent synthetic nanobody neutralizes SARS-CoV-2 by stabilizing inactive Spike.
Science, 370, 2020
|
|
7KKL
 
 | | SARS-CoV-2 Spike in complex with neutralizing nanobody mNb6 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Schoof, M.S, Faust, B.F, Saunders, R.A, Sangwan, S, Rezelj, V, Hoppe, N, Boone, M, Billesboelle, C.B, Puchades, C, Azumaya, C.M, Kratochvil, H.T, Zimanyi, M, Desphande, I, Liang, J, Dickinson, S, Nguyen, H.C, Chio, C.M, Merz, G.E, Thompson, M.C, Diwanji, D, Schaefer, K, Anand, A.A, Dobzinski, N, Zha, B.S, Simoneau, C.R, Leon, K, White, K.M, Chio, U.S, Gupta, M, Jin, M, Li, F, Liu, Y, Zhang, K, Bulkley, D, Sun, M, Smith, A.M, Rizo, A.N, Moss, F, Brilot, A.F, Pourmal, S, Trenker, R, Pospiech, T, Gupta, S, Barsi-Rhyne, B, Belyy, V, Barile-Hill, A.W, Nock, S, Liu, Y, Krogan, N.J, Ralston, C.Y, Swaney, D.L, Garcia-Sastre, A, Ott, M, Vignuzzi, M, Walter, P, Manglik, A, QCRG Structural Biology Consortium | | Deposit date: | 2020-10-27 | | Release date: | 2020-11-11 | | Last modified: | 2021-04-21 | | Method: | ELECTRON MICROSCOPY (2.85 Å) | | Cite: | An ultrapotent synthetic nanobody neutralizes SARS-CoV-2 by stabilizing inactive Spike.
Science, 370, 2020
|
|
5S4J
 
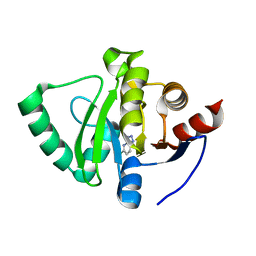 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 Nsp3 macrodomain in complex with SF054 | | Descriptor: | 6-chlorotetrazolo[1,5-b]pyridazine, Non-structural protein 3 | | Authors: | Fearon, D, Schuller, M, Rangel, V.L, Douangamath, A, Rack, J.G.M, Zhu, K, Aimon, A, Brandao-Neto, J, Dias, A, Dunnet, L, Gorrie-Stone, T.J, Powell, A.J, Krojer, T, Skyner, R, Thompson, W, Ahel, I, von Delft, F. | | Deposit date: | 2020-11-02 | | Release date: | 2021-01-13 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.124 Å) | | Cite: | Fragment binding to the Nsp3 macrodomain of SARS-CoV-2 identified through crystallographic screening and computational docking.
Sci Adv, 7, 2021
|
|
5S4I
 
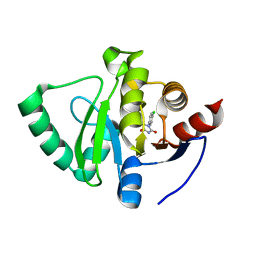 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 Nsp3 macrodomain in complex with SF051 | | Descriptor: | (5S)-1-(4-chlorophenyl)-5-methylimidazolidine-2,4-dione, Non-structural protein 3 | | Authors: | Fearon, D, Schuller, M, Rangel, V.L, Douangamath, A, Rack, J.G.M, Zhu, K, Aimon, A, Brandao-Neto, J, Dias, A, Dunnet, L, Gorrie-Stone, T.J, Powell, A.J, Krojer, T, Skyner, R, Thompson, W, Ahel, I, von Delft, F. | | Deposit date: | 2020-11-02 | | Release date: | 2021-01-13 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.131 Å) | | Cite: | Fragment binding to the Nsp3 macrodomain of SARS-CoV-2 identified through crystallographic screening and computational docking.
Sci Adv, 7, 2021
|
|
5S4H
 
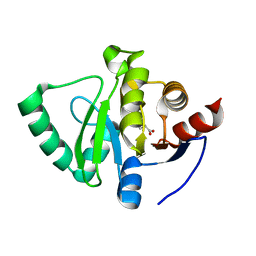 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 Nsp3 macrodomain in complex with SF048 | | Descriptor: | 1-carbamoylpiperidine-4-carboxylic acid, Non-structural protein 3 | | Authors: | Fearon, D, Schuller, M, Rangel, V.L, Douangamath, A, Rack, J.G.M, Zhu, K, Aimon, A, Brandao-Neto, J, Dias, A, Dunnet, L, Gorrie-Stone, T.J, Powell, A.J, Krojer, T, Skyner, R, Thompson, W, Ahel, I, von Delft, F. | | Deposit date: | 2020-11-02 | | Release date: | 2021-01-13 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.175 Å) | | Cite: | Fragment binding to the Nsp3 macrodomain of SARS-CoV-2 identified through crystallographic screening and computational docking.
Sci Adv, 7, 2021
|
|
5S4F
 
 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 Nsp3 macrodomain in complex with SF003 | | Descriptor: | 1,8-naphthyridine, Non-structural protein 3, SULFATE ION | | Authors: | Fearon, D, Schuller, M, Rangel, V.L, Douangamath, A, Rack, J.G.M, Zhu, K, Aimon, A, Brandao-Neto, J, Dias, A, Dunnet, L, Gorrie-Stone, T.J, Powell, A.J, Krojer, T, Skyner, R, Thompson, W, Ahel, I, von Delft, F. | | Deposit date: | 2020-11-02 | | Release date: | 2021-01-13 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.131 Å) | | Cite: | Fragment binding to the Nsp3 macrodomain of SARS-CoV-2 identified through crystallographic screening and computational docking.
Sci Adv, 7, 2021
|
|
5S4G
 
 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 Nsp3 macrodomain in complex with SF005 | | Descriptor: | Non-structural protein 3, SULFATE ION, [1,2,4]triazolo[4,3-a]pyridin-3-amine | | Authors: | Fearon, D, Schuller, M, Rangel, V.L, Douangamath, A, Rack, J.G.M, Zhu, K, Aimon, A, Brandao-Neto, J, Dias, A, Dunnet, L, Gorrie-Stone, T.J, Powell, A.J, Krojer, T, Skyner, R, Thompson, W, Ahel, I, von Delft, F. | | Deposit date: | 2020-11-02 | | Release date: | 2021-01-13 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.172 Å) | | Cite: | Fragment binding to the Nsp3 macrodomain of SARS-CoV-2 identified through crystallographic screening and computational docking.
Sci Adv, 7, 2021
|
|
5S20
 
 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 Nsp3 macrodomain in complex with PB1827975385 | | Descriptor: | (5R)-5-amino-5,6,7,8-tetrahydronaphthalen-1-ol, Non-structural protein 3 | | Authors: | Fearon, D, Schuller, M, Rangel, V.L, Douangamath, A, Rack, J.G.M, Zhu, K, Aimon, A, Brandao-Neto, J, Dias, A, Dunnet, L, Gorrie-Stone, T.J, Powell, A.J, Krojer, T, Skyner, R, Thompson, W, Ahel, I, von Delft, F. | | Deposit date: | 2020-11-02 | | Release date: | 2021-01-13 | | Last modified: | 2022-05-25 | | Method: | X-RAY DIFFRACTION (1.037 Å) | | Cite: | Fragment binding to the Nsp3 macrodomain of SARS-CoV-2 identified through crystallographic screening and computational docking.
Sci Adv, 7, 2021
|
|
