7ALP
 
 | |
1H3N
 
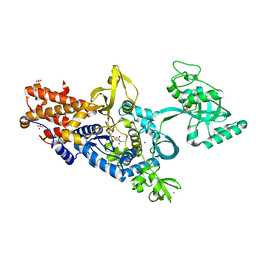 | | Leucyl-tRNA synthetase from Thermus thermophilus complexed with a sulphamoyl analogue of leucyl-adenylate | | Descriptor: | LEUCINE, LEUCYL-TRNA SYNTHETASE, SULFATE ION, ... | | Authors: | Cusack, S, Yaremchuk, A, Tukalo, M. | | Deposit date: | 2002-09-11 | | Release date: | 2002-09-27 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The 2A Structure of Leucyl-tRNA Synthetase and its Complex with a Leucyl-Adenylate Analogue
Embo J., 19, 2000
|
|
1SET
 
 | |
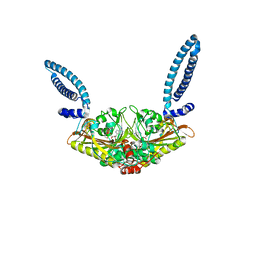
1SES
 
 | |
1OBH
 
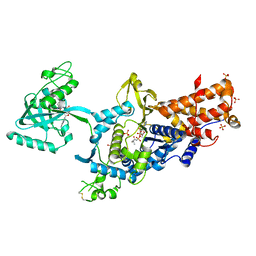 | | LEUCYL-TRNA SYNTHETASE FROM THERMUS THERMOPHILUS COMPLEXED WITH A PRE-TRANSFER EDITING SUBSTRATE ANALOGUE IN BOTH SYNTHETIC ACTIVE SITE AND EDITING SITE | | Descriptor: | LEUCYL-TRNA SYNTHETASE, MERCURY (II) ION, NORVALINE, ... | | Authors: | Cusack, S, Yaremchuk, A, Tukalo, M. | | Deposit date: | 2003-01-31 | | Release date: | 2003-05-09 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural and Mechanistic Basis of Pre- and Posttransfer Editing by Leucyl-tRNA Synthetase
Mol.Cell, 11, 2003
|
|
1OBC
 
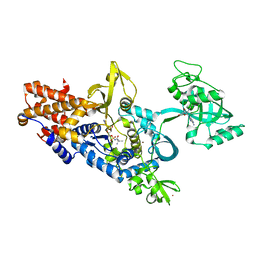 | | LEUCYL-TRNA SYNTHETASE FROM THERMUS THERMOPHILUS COMPLEXED WITH A POST-TRANSFER EDITING SUBSTRATE ANALOGUE | | Descriptor: | 2'-AMINO-2'-DEOXYADENOSINE, LEUCINE, LEUCYL-TRNA SYNTHETASE, ... | | Authors: | Cusack, S, Yaremchuk, A, Tukalo, M. | | Deposit date: | 2003-01-30 | | Release date: | 2003-05-09 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural and Mechanistic Basis of Pre- and Posttransfer Editing by Leucyl-tRNA Synthetase
Mol.Cell, 11, 2003
|
|
1ADJ
 
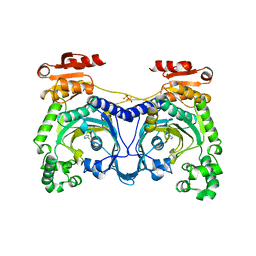 | | HISTIDYL-TRNA SYNTHETASE IN COMPLEX WITH HISTIDINE | | Descriptor: | HISTIDINE, HISTIDYL-TRNA SYNTHETASE, SULFATE ION | | Authors: | Cusack, S, Aberg, A. | | Deposit date: | 1997-02-18 | | Release date: | 1997-08-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure analysis of the activation of histidine by Thermus thermophilus histidyl-tRNA synthetase.
Biochemistry, 36, 1997
|
|
8BEK
 
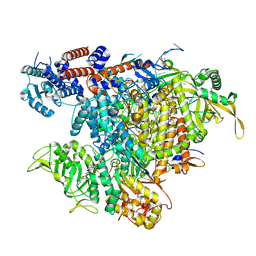 | |
8BE0
 
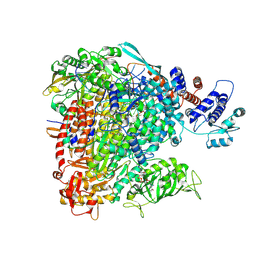 | |
8BF5
 
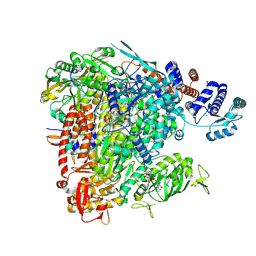 | |
1ADY
 
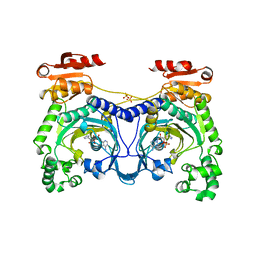 | | HISTIDYL-TRNA SYNTHETASE IN COMPLEX WITH HISTIDYL-ADENYLATE | | Descriptor: | HISTIDYL-ADENOSINE MONOPHOSPHATE, HISTIDYL-TRNA SYNTHETASE, SULFATE ION | | Authors: | Cusack, S, Aberg, A. | | Deposit date: | 1997-02-19 | | Release date: | 1997-08-20 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure analysis of the activation of histidine by Thermus thermophilus histidyl-tRNA synthetase.
Biochemistry, 36, 1997
|
|
6QCX
 
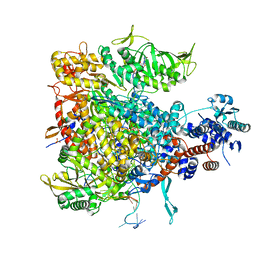 | |
6QCV
 
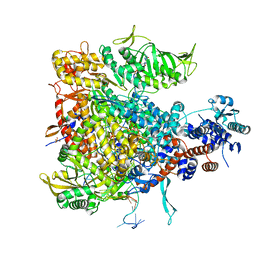 | |
7QTL
 
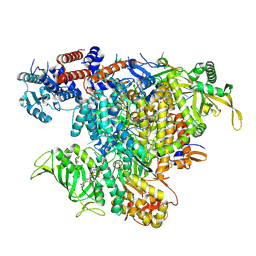 | | Influenza A/H7N9 polymerase elongation complex | | Descriptor: | 3' vRNA, 5' vRNA, 5'-O-[(S)-hydroxy{[(S)-hydroxy(phosphonooxy)phosphoryl]methyl}phosphoryl]cytidine, ... | | Authors: | Cusack, S, Kouba, T. | | Deposit date: | 2022-01-14 | | Release date: | 2022-12-28 | | Last modified: | 2023-02-15 | | Method: | ELECTRON MICROSCOPY (2.48 Å) | | Cite: | Direct observation of backtracking by influenza A and B polymerases upon consecutive incorporation of the nucleoside analog T1106.
Cell Rep, 42, 2023
|
|
7R1F
 
 | |
7R0E
 
 | |
6Z6B
 
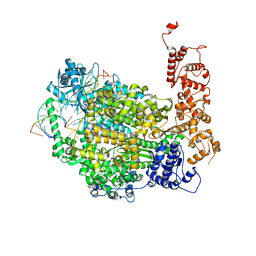 | | Structure of full-length La Crosse virus L protein (polymerase) | | Descriptor: | RNA (5'-R(*GP*CP*UP*AP*CP*UP*AP*A)-3'), RNA (5'-R(P*AP*GP*UP*AP*GP*UP*GP*UP*GP*C)-3'), RNA (5'-R(P*UP*UP*AP*GP*UP*AP*GP*UP*AP*CP*AP*CP*UP*AP*CP*U)-3'), ... | | Authors: | Cusack, S, Gerlach, P, Reguera, J. | | Deposit date: | 2020-05-28 | | Release date: | 2020-07-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.961 Å) | | Cite: | Pre-initiation and elongation structures of full-length La Crosse virus polymerase reveal functionally important conformational changes.
Nat Commun, 11, 2020
|
|
7Z43
 
 | |
7Z4O
 
 | |
7Z42
 
 | |
4WSB
 
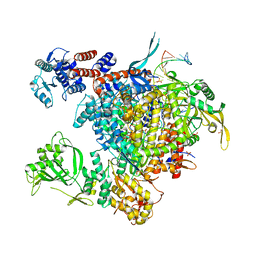 | | Bat Influenza A polymerase with bound vRNA promoter | | Descriptor: | Influenza A polymerase vRNA promoter 3' end, Influenza A polymerase vRNA promoter 5' end, PHOSPHATE ION, ... | | Authors: | Cusack, S, Pflug, A, Guilligay, D, Reich, S. | | Deposit date: | 2014-10-26 | | Release date: | 2014-11-19 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structural insight into cap-snatching and RNA synthesis by influenza polymerase.
Nature, 516, 2014
|
|
5MSG
 
 | |
7ZPL
 
 | |
7ZPM
 
 | | Influenza A/H7N9 polymerase apo-protein dimer complex | | Descriptor: | Polymerase acidic protein, Polymerase basic protein 2, RNA-directed RNA polymerase catalytic subunit | | Authors: | Cusack, S, Kouba, T. | | Deposit date: | 2022-04-27 | | Release date: | 2022-12-28 | | Last modified: | 2023-02-15 | | Method: | ELECTRON MICROSCOPY (2.81 Å) | | Cite: | Direct observation of backtracking by influenza A and B polymerases upon consecutive incorporation of the nucleoside analog T1106.
Cell Rep, 42, 2023
|
|
8BDR
 
 | |
