6HMI
 
 | |
2KO0
 
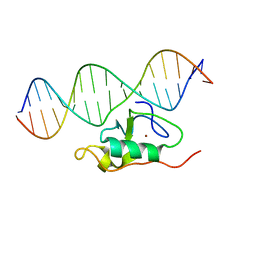 | | Solution structure of the THAP zinc finger of THAP1 in complex with its DNA target | | Descriptor: | RRM1, THAP domain-containing protein 1, ZINC ION | | Authors: | Campagne, S, Gervais, V, Saurel, O, Milon, A. | | Deposit date: | 2009-09-08 | | Release date: | 2010-01-19 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structural determinants of specific DNA-recognition by the THAP zinc finger
Nucleic Acids Res., 2010
|
|
6HMO
 
 | |
5IEJ
 
 | |
5IEB
 
 | |
7ZAP
 
 | |
7Q33
 
 | |
7AEP
 
 | | Solution structure of U1-A RRM2 (190-282) | | Descriptor: | U1 small nuclear ribonucleoprotein A | | Authors: | Campagne, S, Allain, F.H. | | Deposit date: | 2020-09-18 | | Release date: | 2021-02-03 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | An in vitro reconstituted U1 snRNP allows the study of the disordered regions of the particle and the interactions with proteins and ligands.
Nucleic Acids Res., 49, 2021
|
|
4LUP
 
 | | Crystal structure of the complex formed by region of E. coli sigmaE bound to its -10 element non template strand | | Descriptor: | 1,2-ETHANEDIOL, RNA polymerase sigma factor, region 2 of sigmaE of E. coli | | Authors: | Campagne, S, Marsh, M.E, Vorholt, J.A.V, Allain, F.H.-T, Capitani, G. | | Deposit date: | 2013-07-25 | | Release date: | 2014-02-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structural basis for -10 promoter element melting by environmentally induced sigma factors.
Nat.Struct.Mol.Biol., 21, 2014
|
|
2LFW
 
 | | NMR structure of the PhyRSL-NepR complex from Sphingomonas sp. Fr1 | | Descriptor: | NepR anti sigma factor, PhyR sigma-like domain | | Authors: | Campagne, S, Damberger, F.F, Vorholt, J.A, Allain, F.H.-T. | | Deposit date: | 2011-07-18 | | Release date: | 2012-04-25 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural basis for sigma factor mimicry in the general stress response of Alphaproteobacteria.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
2MAO
 
 | |
2MAP
 
 | |
2L1G
 
 | | RDC refined solution structure of the THAP zinc finger of THAP1 in complex with its 16bp RRM1 DNA target | | Descriptor: | DNA (5'-D(*GP*CP*TP*TP*GP*TP*GP*TP*GP*GP*GP*CP*AP*GP*CP*G)-3'), DNA (5'-D(P*CP*GP*CP*TP*GP*CP*CP*CP*AP*CP*AP*CP*AP*AP*GP*C)-3'), THAP domain-containing protein 1, ... | | Authors: | Campagne, S, Gervais, V, Saurel, O, Milon, A. | | Deposit date: | 2010-07-28 | | Release date: | 2010-09-08 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | RDC refined solution structure of the THAP zinc finger of THAP1 in complex with its 16bp RRM1 DNA target
To be published
|
|
2JOS
 
 | |
5OR5
 
 | |
6SNJ
 
 | |
2NB9
 
 | | Solution structure of ZitP zinc finger | | Descriptor: | Uncharacterized protein, ZINC ION | | Authors: | Campagne, S, Berge, M, Viollier, P.H, Allain, F.H.-T. | | Deposit date: | 2016-02-01 | | Release date: | 2016-12-14 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Modularity and determinants of a (bi-)polarization control system from free-living and obligate intracellular bacteria.
Elife, 5, 2016
|
|
5OR0
 
 | |
3BEW
 
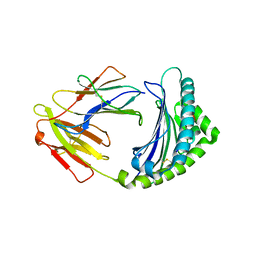 | | 10mer Crystal Structure of chicken MHC class I haplotype B21 | | Descriptor: | 10-mer from Tubulin beta-6 chain, Beta-2-microglobulin, Major histocompatibility complex class I glycoprotein haplotype B21 | | Authors: | Koch, M, Camp, S, Collen, T, Avila, D, Salomonsen, J, Wallny, H.J, van Hateren, A, Hunt, L, Jacob, J.P, Johnston, F, Marston, D.A, Shaw, I, Dunbar, P.R, Cerundolo, V, Jones, E.Y, Kaufman, J. | | Deposit date: | 2007-11-20 | | Release date: | 2008-01-01 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structures of an MHC class I molecule from b21 chickens illustrate promiscuous Peptide binding
Immunity, 27, 2007
|
|
8CF2
 
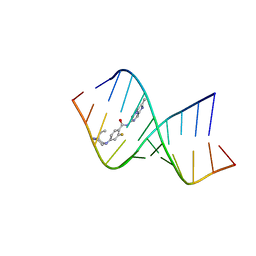 | | Solution structure of the RNA helix formed by the 5'-end of U1 snRNA and an A-1 bulged 5'-splice site in complex with SMN-CY | | Descriptor: | 4-[(3~{S})-3-ethylpiperazin-1-yl]-2-fluoranyl-~{N}-(2-methylimidazo[1,2-a]pyrazin-6-yl)benzamide, RNA (5'-R(P*AP*UP*AP*CP*(PSU)P*(PSU)P*AP*CP*CP*UP*G)-3'), RNA (5'-R(P*GP*GP*AP*GP*UP*AP*AP*GP*UP*CP*U)-3') | | Authors: | Malard, F, Marquevielle, J, Campagne, S. | | Deposit date: | 2023-02-02 | | Release date: | 2024-02-21 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The diversity of splicing modifiers acting on A-1 bulged 5'-splice sites reveals rules for rational drug design.
Nucleic Acids Res., 52, 2024
|
|
6EZ6
 
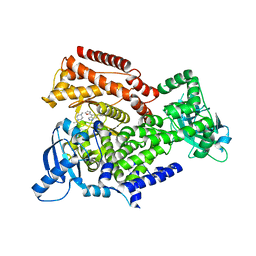 | | PI3 kinase delta in complex with Methyl 5-(4-(5-((4-isopropylpiperazin-1-yl)methyl)oxazol-2-yl)-1H-indazol-6-yl)-2-methoxynicotinate | | Descriptor: | Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit delta isoform, methyl 2-methoxy-5-[4-[5-[(4-propan-2-ylpiperazin-1-yl)methyl]-1,3-oxazol-2-yl]-2~{H}-indazol-6-yl]pyridine-3-carboxylate | | Authors: | Convery, M.A, Campos, S, Dalton, S.E. | | Deposit date: | 2017-11-14 | | Release date: | 2017-12-27 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Selectively Targeting the Kinome-Conserved Lysine of PI3K delta as a General Approach to Covalent Kinase Inhibition.
J. Am. Chem. Soc., 140, 2018
|
|
6EYZ
 
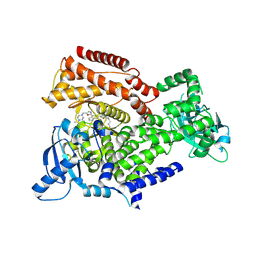 | | PI3 kinase delta in complex with 4-Fluorophenyl 5-(4-(5-((4-isopropylpiperazin-1-yl)methyl)oxazol-2-yl)-1H-indazol-6-yl)-2-methoxynicotinate | | Descriptor: | 2-methoxy-5-[4-[5-[(4-propan-2-ylpiperazin-1-yl)methyl]-1,3-oxazol-2-yl]-2~{H}-indazol-6-yl]pyridine-3-carboxylic acid, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit delta isoform | | Authors: | Convery, M.A, Campos, S, Dalton, S.E. | | Deposit date: | 2017-11-13 | | Release date: | 2017-12-20 | | Last modified: | 2018-01-31 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Selectively Targeting the Kinome-Conserved Lysine of PI3K delta as a General Approach to Covalent Kinase Inhibition.
J. Am. Chem. Soc., 140, 2018
|
|
8R8P
 
 | |
8R62
 
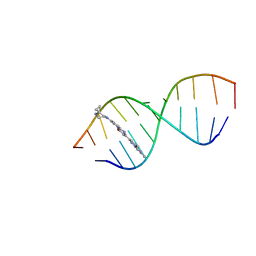 | | Solution structure of Risdiplam bound to the RNA duplex formed upon 5'-splice site recognition | | Descriptor: | 7-(4,7-diazaspiro[2.5]octan-7-yl)-2-(2,8-dimethylimidazo[1,2-b]pyridazin-6-yl)-1~{H}-pyrido[1,2-a]pyrimidin-4-one, RNA (5'-R(*AP*UP*AP*CP*(PSU)P*(PSU)P*AP*CP*CP*UP*G)-3'), RNA (5'-R(P*GP*GP*AP*GP*UP*AP*AP*GP*UP*CP*U)-3') | | Authors: | Malard, F, Campagne, S. | | Deposit date: | 2023-11-20 | | Release date: | 2024-03-06 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The diversity of splicing modifiers acting on A-1 bulged 5'-splice sites reveals rules for rational drug design.
Nucleic Acids Res., 52, 2024
|
|
8R63
 
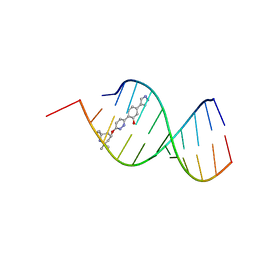 | | Solution structure of branaplam bound to the RNA duplex formed upon 5'-splice site recognition | | Descriptor: | 5-(1~{H}-pyrazol-4-yl)-2-[6-(2,2,6,6-tetramethylpiperidin-4-yl)oxypyridazin-3-yl]phenol, RNA (5'-R(*AP*UP*AP*CP*(PSU)P*(PSU)P*AP*CP*CP*UP*G)-3'), RNA (5'-R(P*GP*GP*AP*GP*UP*AP*AP*GP*UP*CP*U)-3') | | Authors: | Malard, F, Campagne, S. | | Deposit date: | 2023-11-20 | | Release date: | 2024-03-06 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The diversity of splicing modifiers acting on A-1 bulged 5'-splice sites reveals rules for rational drug design.
Nucleic Acids Res., 52, 2024
|
|
