5A63
 
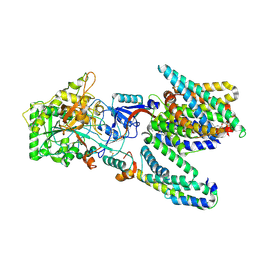 | | Cryo-EM structure of the human gamma-secretase complex at 3.4 angstrom resolution. | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Bai, X, Yan, C, Yang, G, Lu, P, Ma, D, Sun, L, Zhou, R, Scheres, S.H.W, Shi, Y. | | Deposit date: | 2015-06-24 | | Release date: | 2015-08-05 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | An Atomic Structure of Human Gamma-Secretase
Nature, 525, 2015
|
|
3Q0K
 
 | | Crystal structure of Human PACSIN 2 F-BAR | | Descriptor: | CALCIUM ION, Protein kinase C and casein kinase substrate in neurons protein 2 | | Authors: | Bai, X, Meng, G, Zheng, X. | | Deposit date: | 2010-12-15 | | Release date: | 2012-02-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of human PACSIN 2 F-BAR domain
To be Published
|
|
3SYV
 
 | | Crystal structure of mPACSIN 3 F-BAR domain mutant | | Descriptor: | Protein kinase C and casein kinase II substrate protein 3 | | Authors: | Bai, X. | | Deposit date: | 2011-07-18 | | Release date: | 2012-05-16 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Rigidity of wedge loop in PACSIN 3 protein is a key factor in dictating diameters of tubules
J.Biol.Chem., 287, 2012
|
|
2OT5
 
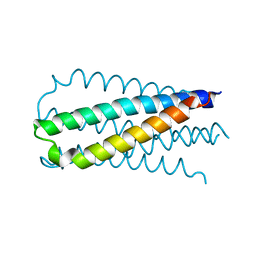 | |
3Q84
 
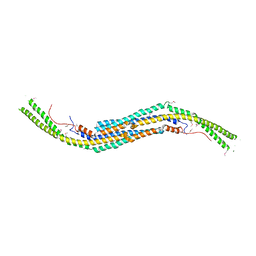 | | Crystal structure of human PACSIN 1 F-BAR domain | | Descriptor: | CALCIUM ION, Protein kinase C and casein kinase substrate in neurons protein 1 | | Authors: | Bai, X. | | Deposit date: | 2011-01-06 | | Release date: | 2012-02-22 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of human PACSIN 1 F-BAR domain
To be Published
|
|
8K9H
 
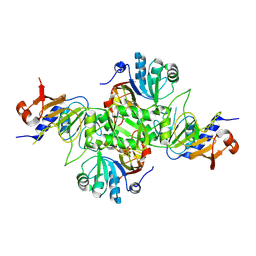 | |
8I29
 
 | | Crystal structure of butanol dehydrogenase A (YqdH) in complex with NADH from Fusobacterium nucleatum | | Descriptor: | CHLORIDE ION, COBALT (II) ION, NADH-dependent butanol dehydrogenase A, ... | | Authors: | Bai, X, Lan, J, Bu, T.T, Wang, L.L, He, S.R, Zhang, J, Xu, Y.B. | | Deposit date: | 2023-01-14 | | Release date: | 2023-02-01 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.72 Å) | | Cite: | Crystal structure of butanol dehydrogenase A (YqdH) in complex with NADH from Fusobacterium nucleatum
To Be Published
|
|
7FJG
 
 | | Crystal structure of butanol dehydrogenase A (YqdH) in complex with partial NADH from Fusobacterium nucleatum | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, FE (III) ION, NADH-dependent butanol dehydrogenase A | | Authors: | Bai, X, Lan, J, Wang, L, Bu, T, Xu, Y. | | Deposit date: | 2021-08-03 | | Release date: | 2022-06-15 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.72 Å) | | Cite: | Crystal structure of butanol dehydrogenase A (YqdH) in complex with partial NADH from Fusobacterium nucleatum
To Be Published
|
|
3QE6
 
 | | Mouse PACSIN 3 F-BAR domain structure | | Descriptor: | MAGNESIUM ION, Protein kinase C and casein kinase II substrate protein 3 | | Authors: | Meng, G, Bai, X, Zheng, X. | | Deposit date: | 2011-01-19 | | Release date: | 2012-01-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Rigidity of wedge loop in PACSIN 3 protein is a key factor in dictating diameters of tubules
J.Biol.Chem., 287, 2012
|
|
8HGS
 
 | | The EGF-bound EGFR ectodomain homodimer | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Epidermal growth factor receptor, Pro-epidermal growth factor, ... | | Authors: | Zhang, Z, Bai, X. | | Deposit date: | 2022-11-15 | | Release date: | 2023-02-15 | | Last modified: | 2023-03-01 | | Method: | ELECTRON MICROSCOPY (3.81 Å) | | Cite: | Structure and dynamics of the EGFR/HER2 heterodimer.
Cell Discov, 9, 2023
|
|
8HGO
 
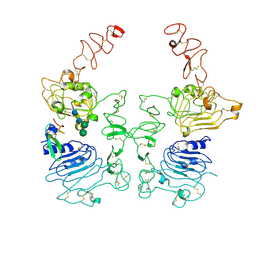 | | The EGF-bound EGFR/HER2 ectodomain complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Epidermal growth factor, ... | | Authors: | Zhang, Z, Bai, X. | | Deposit date: | 2022-11-15 | | Release date: | 2023-02-08 | | Last modified: | 2023-03-01 | | Method: | ELECTRON MICROSCOPY (3.31 Å) | | Cite: | Structure and dynamics of the EGFR/HER2 heterodimer.
Cell Discov, 9, 2023
|
|
8HGP
 
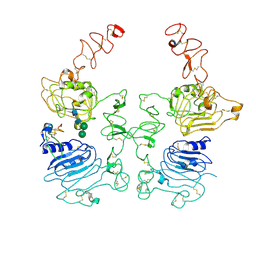 | | The EREG-bound EGFR/HER2 ectodomain complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Epidermal growth factor receptor, ... | | Authors: | Zhang, Z, Bai, X. | | Deposit date: | 2022-11-15 | | Release date: | 2023-02-08 | | Last modified: | 2023-03-01 | | Method: | ELECTRON MICROSCOPY (4.53 Å) | | Cite: | Structure and dynamics of the EGFR/HER2 heterodimer.
Cell Discov, 9, 2023
|
|
4V92
 
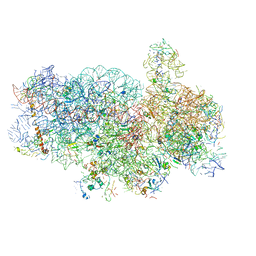 | | Kluyveromyces lactis 80S ribosome in complex with CrPV-IRES | | Descriptor: | 18S RRNA, ES1, ES10, ... | | Authors: | Fernandez, I.S, Bai, X, Scheres, S.H.W, Ramakrishnan, V. | | Deposit date: | 2014-03-21 | | Release date: | 2014-07-09 | | Last modified: | 2019-12-11 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Initiation of Translation by Cricket Paralysis Virus Ires Requires its Translocation in the Ribosome.
Cell(Cambridge,Mass.), 157, 2014
|
|
4V91
 
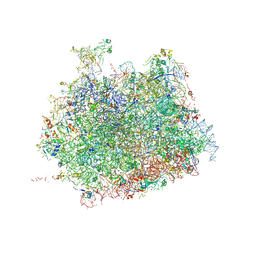 | | Kluyveromyces lactis 80S ribosome in complex with CrPV-IRES | | Descriptor: | 25S RRNA, 5.8S RRNA, 5S RRNA, ... | | Authors: | Fernandez, I.S, Bai, X, Scheres, S.H.W, Ramakrishnan, V. | | Deposit date: | 2014-03-21 | | Release date: | 2014-07-09 | | Last modified: | 2019-12-11 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Initiation of Translation by Cricket Paralysis Virus Ires Requires its Translocation in the Ribosome.
Cell(Cambridge,Mass.), 157, 2014
|
|
4AYD
 
 | | Structure of a complex between CCPs 6 and 7 of Human Complement Factor H and Neisseria meningitidis FHbp Variant 1 R106A mutant | | Descriptor: | 1,2-ETHANEDIOL, COMPLEMENT FACTOR H, FACTOR H BINDING PROTEIN | | Authors: | Johnson, S, Tan, L, van der Veen, S, Caesar, J, Goicoechea De Jorge, E, Everett, R.J, Bai, X, Exley, R.M, Ward, P.N, Ruivo, N, Trivedi, K, Cumber, E, Jones, R, Newham, L, Staunton, D, Borrow, R, Pickering, M, Lea, S.M, Tang, C.M. | | Deposit date: | 2012-06-20 | | Release date: | 2012-11-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Design and Evaluation of Meningococcal Vaccines Through Structure-Based Modification of Host and Pathogen Molecules
Plos Pathog., 8, 2012
|
|
4AYM
 
 | | Structure of a complex between CCPs 6 and 7 of Human Complement Factor H and Neisseria meningitidis FHbp Variant 3 P106A mutant | | Descriptor: | COMPLEMENT FACTOR H, FACTOR H BINDING PROTEIN | | Authors: | Johnson, S, Tan, L, van der Veen, S, Caesar, J, Goicoechea De Jorge, E, Everett, R.J, Bai, X, Exley, R.M, Ward, P.N, Ruivo, N, Trivedi, K, Cumber, E, Jones, R, Newham, L, Staunton, D, Borrow, R, Pickering, M, Lea, S.M, Tang, C.M. | | Deposit date: | 2012-06-21 | | Release date: | 2012-11-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Design and Evaluation of Meningococcal Vaccines Through Structure-Based Modification of Host and Pathogen Molecules.
Plos Pathog., 8, 2012
|
|
4AYE
 
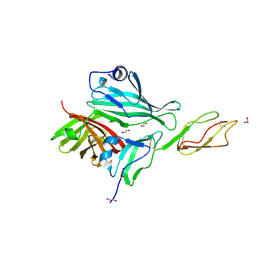 | | Structure of a complex between CCPs 6 and 7 of Human Complement Factor H and Neisseria meningitidis FHbp Variant 1 E283AE304A mutant | | Descriptor: | 1,2-ETHANEDIOL, COMPLEMENT FACTOR H, FACTOR H BINDING PROTEIN | | Authors: | Johnson, S, Tan, L, van der Veen, S, Caesar, J, Goicoechea De Jorge, E, Everett, R.J, Bai, X, Exley, R.M, Ward, P.N, Ruivo, N, Trivedi, K, Cumber, E, Jones, R, Newham, L, Staunton, D, Borrow, R, Pickering, M, Lea, S.M, Tang, C.M. | | Deposit date: | 2012-06-20 | | Release date: | 2012-11-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Design and Evaluation of Meningococcal Vaccines Through Structure-Based Modification of Host and Pathogen Molecules.
Plos Pathog., 8, 2012
|
|
4AYI
 
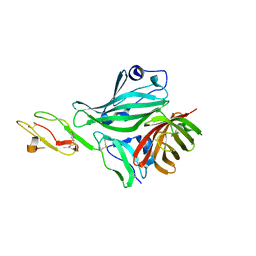 | | Structure of a complex between CCPs 6 and 7 of Human Complement Factor H and Neisseria meningitidis FHbp Variant 3 Wild type | | Descriptor: | 1,2-ETHANEDIOL, COMPLEMENT FACTOR H, LIPOPROTEIN GNA1870 CCOMPND 7 | | Authors: | Johnson, S, Tan, L, van der Veen, S, Caesar, J, Goicoechea De Jorge, E, Everett, R.J, Bai, X, Exley, R.M, Ward, P.N, Ruivo, N, Trivedi, K, Cumber, E, Jones, R, Newham, L, Staunton, D, Borrow, R, Pickering, M, Lea, S.M, Tang, C.M. | | Deposit date: | 2012-06-21 | | Release date: | 2012-11-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Design and Evaluation of Meningococcal Vaccines Through Structure-Based Modification of Host and Pathogen Molecules.
Plos Pathog., 8, 2012
|
|
4AYN
 
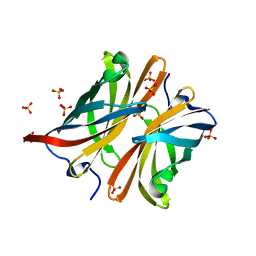 | | Structure of the C-terminal barrel of Neisseria meningitidis FHbp Variant 2 | | Descriptor: | FACTOR H-BINDING PROTEIN, SULFATE ION | | Authors: | Johnson, S, Tan, L, van der Veen, S, Caesar, J, Goicoechea De Jorge, E, Everett, R.J, Bai, X, Exley, R.M, Ward, P.N, Ruivo, N, Trivedi, K, Cumber, E, Jones, R, Newham, L, Staunton, D, Borrow, R, Pickering, M, Lea, S.M, Tang, C.M. | | Deposit date: | 2012-06-21 | | Release date: | 2012-11-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Design and Evaluation of Meningococcal Vaccines Through Structure-Based Modification of Host and Pathogen Molecules.
Plos Pathog., 8, 2012
|
|
3QNI
 
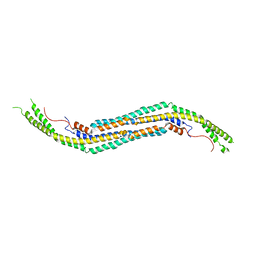 | |
6WPK
 
 | |
6NQ1
 
 | | Cryo-EM structure of human TPC2 channel in the apo state | | Descriptor: | Two pore calcium channel protein 2 | | Authors: | She, J, Zeng, W, Guo, J, Chen, Q, Bai, X, Jiang, Y. | | Deposit date: | 2019-01-18 | | Release date: | 2019-03-27 | | Last modified: | 2019-11-20 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural mechanisms of phospholipid activation of the human TPC2 channel.
Elife, 8, 2019
|
|
6NQ0
 
 | | Cryo-EM structure of human TPC2 channel in the ligand-bound open state | | Descriptor: | (2R)-3-{[(S)-hydroxy{[(1S,2R,3R,4S,5S,6R)-2,4,6-trihydroxy-3,5-bis(phosphonooxy)cyclohexyl]oxy}phosphoryl]oxy}propane-1,2-diyl dioctanoate, Two pore calcium channel protein 2 | | Authors: | She, J, Zeng, W, Guo, J, Chen, Q, Bai, X, Jiang, Y. | | Deposit date: | 2019-01-18 | | Release date: | 2019-03-27 | | Last modified: | 2019-11-20 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural mechanisms of phospholipid activation of the human TPC2 channel.
Elife, 8, 2019
|
|
6NQ2
 
 | | Cryo-EM structure of human TPC2 channel in the ligand-bound closed state | | Descriptor: | (2R)-3-{[(S)-hydroxy{[(1S,2R,3R,4S,5S,6R)-2,4,6-trihydroxy-3,5-bis(phosphonooxy)cyclohexyl]oxy}phosphoryl]oxy}propane-1,2-diyl dioctanoate, Two pore calcium channel protein 2 | | Authors: | She, J, Zeng, W, Guo, J, Chen, Q, Bai, X, Jiang, Y. | | Deposit date: | 2019-01-18 | | Release date: | 2019-03-27 | | Last modified: | 2019-11-20 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural mechanisms of phospholipid activation of the human TPC2 channel.
Elife, 8, 2019
|
|
8K1C
 
 | |
