6PIK
 
 | | Tetrameric cryo-EM ArnA | | Descriptor: | Bifunctional polymyxin resistance protein ArnA, UDP-4-amino-4-deoxy-L-arabinose formyltransferase | | Authors: | Yang, M, Gehring, K. | | Deposit date: | 2019-06-26 | | Release date: | 2019-07-31 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (7.8 Å) | | Cite: | Cryo-electron microscopy structures of ArnA, a key enzyme for polymyxin resistance, revealed unexpected oligomerizations and domain movements.
J.Struct.Biol., 208, 2019
|
|
2UXN
 
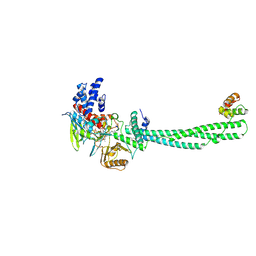 | | Structural Basis of Histone Demethylation by LSD1 Revealed by Suicide Inactivation | | Descriptor: | CHLORIDE ION, DIHYDROFLAVINE-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | Authors: | Yang, M, Culhane, J.C, Szewczuk, L.M, Gocke, C.B, Brautigam, C.A, Tomchick, D.R, Machius, M, Cole, P.A, Yu, H. | | Deposit date: | 2007-03-28 | | Release date: | 2007-05-29 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.72 Å) | | Cite: | Structural Basis of Histone Demethylation by Lsd1 Revealed by Suicide Inactivation.
Nat.Struct.Mol.Biol., 14, 2007
|
|
2UXX
 
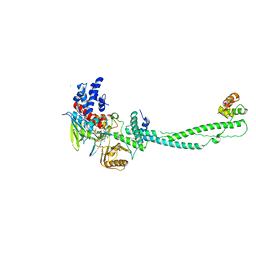 | | Human LSD1 Histone Demethylase-CoREST in complex with an FAD- tranylcypromine adduct | | Descriptor: | CHLORIDE ION, FAD-trans-2-Phenylcyclopropylamine Adduct, GLYCEROL, ... | | Authors: | Yang, M, Culhane, J.C, Machius, M, Cole, P.A, Yu, H. | | Deposit date: | 2007-03-30 | | Release date: | 2007-08-21 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.74 Å) | | Cite: | Structural Basis for the Inhibition of the Lsd1 Histone Demethylase by the Antidepressant Trans-2-Phenylcyclopropylamine.
Biochemistry, 46, 2007
|
|
6PIH
 
 | | Hexameric ArnA cryo-EM structure | | Descriptor: | Bifunctional polymyxin resistance protein ArnA, UDP-4-amino-4-deoxy-L-arabinose formyltransferase | | Authors: | Yang, M, Gehring, K. | | Deposit date: | 2019-06-26 | | Release date: | 2019-07-31 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (6.6 Å) | | Cite: | Cryo-electron microscopy structures of ArnA, a key enzyme for polymyxin resistance, revealed unexpected oligomerizations and domain movements.
J.Struct.Biol., 208, 2019
|
|
2VFX
 
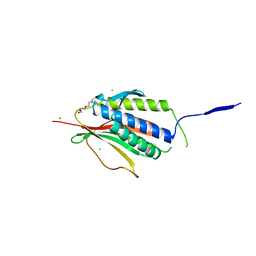 | | Structure of the Symmetric Mad2 Dimer | | Descriptor: | 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, 3,6,9,12,15,18,21,24,27,30,33,36,39-TRIDECAOXAHENTETRACONTANE-1,41-DIOL, CHLORIDE ION, ... | | Authors: | Yang, M, Li, B, Liu, C.-J, Tomchick, D.R, Machius, M, Rizo, J, Yu, H, Luo, X. | | Deposit date: | 2007-11-05 | | Release date: | 2008-03-18 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Insights Into MAD2 Regulation in the Spindle Checkpoint Revealed by the Crystal Structure of the Symmetric MAD2 Dimer.
Plos Biol., 6, 2008
|
|
2IW5
 
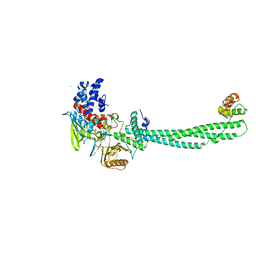 | | Structural Basis for CoREST-Dependent Demethylation of Nucleosomes by the Human LSD1 Histone Demethylase | | Descriptor: | AMMONIUM ION, CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Yang, M, Gocke, C.B, Luo, X, Borek, D, Tomchick, D.R, Machius, M, Otwinowski, Z, Yu, H. | | Deposit date: | 2006-06-26 | | Release date: | 2006-08-09 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | Structural Basis for Corest-Dependent Demethylation of Nucleosomes by the Human Lsd1 Histone Demethylase
Mol.Cell, 23, 2006
|
|
2H2N
 
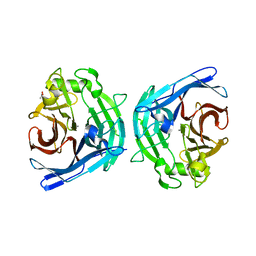 | | Crystal structure of human soluble calcium-activated nucleotidase (SCAN) with calcium ion | | Descriptor: | ACETATE ION, CALCIUM ION, Soluble calcium-activated nucleotidase 1 | | Authors: | Yang, M, Horii, K, Herr, A.B, Kirley, T.L. | | Deposit date: | 2006-05-19 | | Release date: | 2006-07-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Calcium-dependent dimerization of human soluble calcium activated nucleotidase: characterization of the dimer interface.
J.Biol.Chem., 281, 2006
|
|
2H2U
 
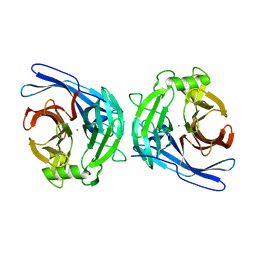 | | Crystal structure of the E130Y mutant of human soluble calcium-activated nucleotidase (SCAN) with calcium ion | | Descriptor: | CALCIUM ION, Soluble calcium-activated nucleotidase 1 | | Authors: | Yang, M, Horii, K, Herr, A.B, Kirley, T.L. | | Deposit date: | 2006-05-19 | | Release date: | 2006-07-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Calcium-dependent dimerization of human soluble calcium activated nucleotidase: characterization of the dimer interface.
J.Biol.Chem., 281, 2006
|
|
4HY6
 
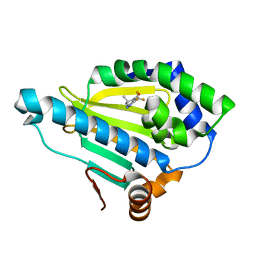 | | Crystal Structure of the human Hsp90-alpha N-domain bound to the hsp90 inhibitor FJ1 | | Descriptor: | 6,6-dimethyl-3-(trifluoromethyl)-1,5,6,7-tetrahydro-4H-indazol-4-one, Heat shock protein HSP 90-alpha | | Authors: | Yang, M, Li, J, He, J.H. | | Deposit date: | 2012-11-13 | | Release date: | 2013-05-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.649 Å) | | Cite: | Crystal Structure of the human Hsp90-alpha N-domain bound to the hsp90 inhibitor FJ1
to be published
|
|
7YDW
 
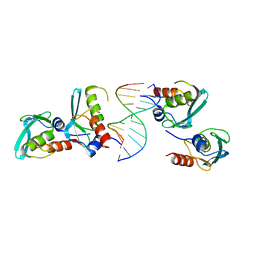 | | Crystal structure of the MPND-DNA complex | | Descriptor: | DNA (5'-D(P*AP*AP*AP*AP*AP*AP*AP*AP*AP*A)-3'), DNA (5'-D(P*TP*TP*TP*TP*TP*TP*TP*TP*TP*T)-3'), MPN domain-containing protein | | Authors: | Yang, M, Chen, Z. | | Deposit date: | 2022-07-04 | | Release date: | 2023-02-15 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | Structures of MPND Reveal the Molecular Recognition of Nucleosomes.
Int J Mol Sci, 24, 2023
|
|
7YDT
 
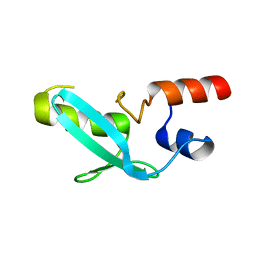 | | Crystal structure of mouse MPND | | Descriptor: | MPN domain containing protein | | Authors: | Yang, M, Chen, Z. | | Deposit date: | 2022-07-04 | | Release date: | 2023-02-15 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.055 Å) | | Cite: | Structures of MPND Reveal the Molecular Recognition of Nucleosomes.
Int J Mol Sci, 24, 2023
|
|
5JYK
 
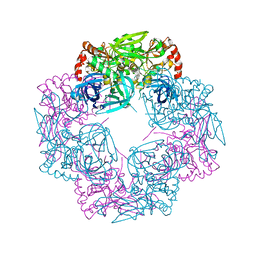 | | Deg9 crystal under 289K | | Descriptor: | GLYCEROL, Protease Do-like 9 | | Authors: | Ouyang, M, Zhang, L.X. | | Deposit date: | 2016-05-14 | | Release date: | 2017-05-31 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.297 Å) | | Cite: | The crystal structure of Deg9 reveals a novel octameric-type HtrA protease
Nat Plants, 3, 2017
|
|
5ILA
 
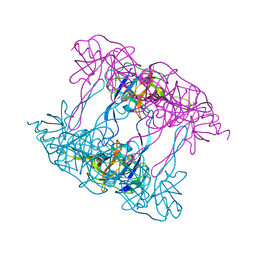 | | Deg9 protease domain | | Descriptor: | Protease Do-like 9 | | Authors: | Ouyang, M, Liu, L, Li, X.Y, Zhao, S, Zhang, L.X. | | Deposit date: | 2016-03-04 | | Release date: | 2017-03-08 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.001 Å) | | Cite: | The crystal structure of Deg9 reveals a novel octameric-type HtrA protease
Nat Plants, 3, 2017
|
|
5ILB
 
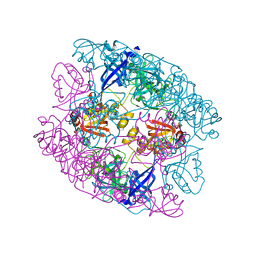 | | Crystal structure of protease domain of Deg2 linked with the PDZ domain of Deg9 | | Descriptor: | Protease Do-like 2, chloroplastic,Protease Do-like 9 | | Authors: | Ouyang, M, Liu, L, Li, X.Y, Zhao, S, Zhang, L.X. | | Deposit date: | 2016-03-04 | | Release date: | 2017-03-08 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.852 Å) | | Cite: | The crystal structure of Deg9 reveals a novel octameric-type HtrA protease
Nat Plants, 3, 2017
|
|
5IL9
 
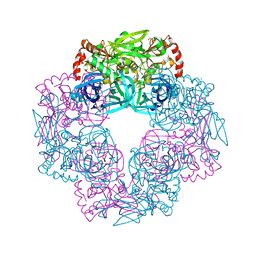 | | Crystal structure of Deg9 | | Descriptor: | GLYCEROL, Protease Do-like 9 | | Authors: | Ouyang, M, Liu, L, Li, X.Y, Zhao, S, Zhang, L.X. | | Deposit date: | 2016-03-04 | | Release date: | 2017-03-08 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The crystal structure of Deg9 reveals a novel octameric-type HtrA protease
Nat Plants, 3, 2017
|
|
5Y09
 
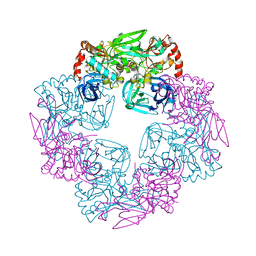 | |
6JD6
 
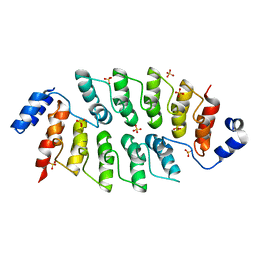 | |
4W78
 
 | | Crystal structure of the ChsH1-ChsH2 complex from Mycobacterium tuberculosis | | Descriptor: | CADMIUM ION, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Guja, K.E, Yang, M, Sampson, N, Garcia-Diaz, M. | | Deposit date: | 2014-08-21 | | Release date: | 2014-10-08 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.541 Å) | | Cite: | A Distinct MaoC-like Enoyl-CoA Hydratase Architecture Mediates Cholesterol Catabolism in Mycobacterium tuberculosis.
Acs Chem.Biol., 9, 2014
|
|
4WNB
 
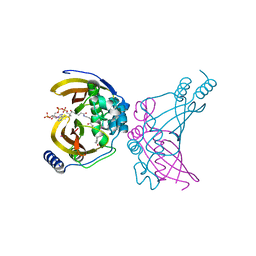 | | Crystal structure of the ChsH1-ChsH2 complex from Mycobacterium tuberculosis bound to 3-OPC-CoA | | Descriptor: | 3-oxo-4-pregnene-20-carboxyl-Coenzyme A, CADMIUM ION, CALCIUM ION, ... | | Authors: | Guja, K.E, Yang, M, Sampson, N, Garcia-Diaz, M. | | Deposit date: | 2014-10-11 | | Release date: | 2014-10-29 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | A Distinct MaoC-like Enoyl-CoA Hydratase Architecture Mediates Cholesterol Catabolism in Mycobacterium tuberculosis.
Acs Chem.Biol., 9, 2014
|
|
4X28
 
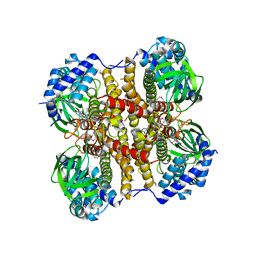 | | Crystal structure of the ChsE4-ChsE5 complex from Mycobacterium tuberculosis | | Descriptor: | Acyl-CoA dehydrogenase, DIHYDROFLAVINE-ADENINE DINUCLEOTIDE | | Authors: | Guja, K.E, Yang, M, Sampson, N, Garcia-Diaz, M. | | Deposit date: | 2014-11-26 | | Release date: | 2015-02-18 | | Last modified: | 2019-12-18 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Unraveling Cholesterol Catabolism in Mycobacterium tuberculosis: ChsE4-ChsE5 alpha 2 beta 2 Acyl-CoA Dehydrogenase Initiates beta-Oxidation of 3-Oxo-cholest-4-en-26-oyl CoA.
Acs Infect Dis., 1, 2015
|
|
5I5N
 
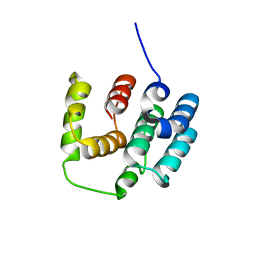 | |
5I5O
 
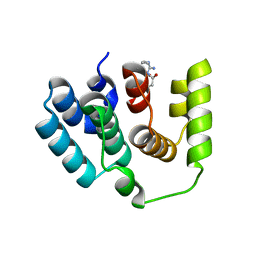 | |
4IKG
 
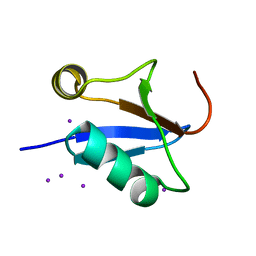 | |
4NQJ
 
 | | Structure of coiled-coil domain | | Descriptor: | DODECYL-BETA-D-MALTOSIDE, E3 ubiquitin-protein ligase TRIM69 | | Authors: | Yang, M, Li, Y. | | Deposit date: | 2013-11-25 | | Release date: | 2014-05-21 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.152 Å) | | Cite: | Structural insights into the TRIM family of ubiquitin E3 ligases.
Cell Res., 24, 2014
|
|
3T9N
 
 | | Crystal structure of a membrane protein | | Descriptor: | DODECYL-BETA-D-MALTOSIDE, Small-conductance mechanosensitive channel | | Authors: | Yang, M, Zhang, X, Ge, J, Wang, J. | | Deposit date: | 2011-08-03 | | Release date: | 2012-10-31 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.456 Å) | | Cite: | Structure and molecular mechanism of an anion-selective mechanosensitive channel of small conductance
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
