1QZ2
 
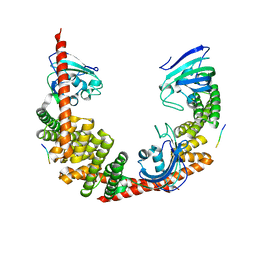 | | Crystal Structure of FKBP52 C-terminal Domain complex with the C-terminal peptide MEEVD of Hsp90 | | Descriptor: | 5-mer peptide from Heat shock protein HSP 90, FK506-binding protein 4 | | Authors: | Wu, B, Li, P, Lou, Z, Liu, Y, Ding, Y, Shu, C, Shen, B, Rao, Z. | | Deposit date: | 2003-09-15 | | Release date: | 2004-06-22 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | 3D structure of human FK506-binding protein 52: Implications for the assembly of the glucocorticoid receptor/Hsp90/immunophilin heterocomplex.
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
3REH
 
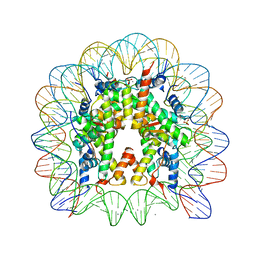 | |
2KKU
 
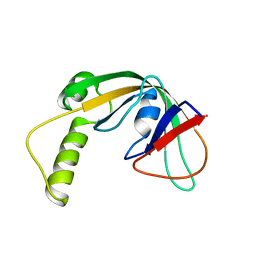 | | Solution structure of protein af2351 from Archaeoglobus fulgidus. Northeast Structural Genomics Consortium target AtT9/Ontario Center for Structural Proteomics Target af2351 | | Descriptor: | Uncharacterized protein | | Authors: | Wu, B, Yee, A, Fares, C, Lemak, A, Rumpel, S, Semest, A, Montelione, G.T, Arrowsmith, C.H, Northeast Structural Genomics Consortium (NESG), Ontario Centre for Structural Proteomics (OCSP) | | Deposit date: | 2009-06-29 | | Release date: | 2009-07-14 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Solution structure of protein af2351 from Archaeoglobus fulgidus. Northeast Structural Genomics Consortium target AtT9/Ontario Center for Structural Proteomics Target af2351
To be Published
|
|
2KQ9
 
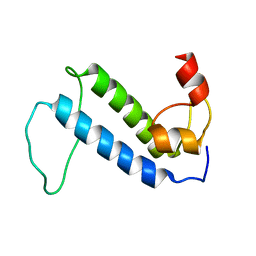 | | Solution structure of DnaK suppressor protein from Agrobacterium tumefaciens C58. Northeast Structural Genomics Consortium target AtT12/Ontario Center for Structural Proteomics Target atc0888 | | Descriptor: | DnaK suppressor protein, ZINC ION | | Authors: | Wu, B, Yee, A, Fares, C, Lemak, A, Semest, A, Montelione, G.T, Arrowsmith, C, Northeast Structural Genomics Consortium (NESG), Ontario Centre for Structural Proteomics (OCSP) | | Deposit date: | 2009-11-02 | | Release date: | 2009-11-17 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of DnaK protein from Agrobacterium tumefaciens C58. Northeast Structural Genomics Consortium target AtT12/Ontario Center for Structural Proteomics Target atc0888
To be Published
|
|
2KYR
 
 | | Solution structure of Enzyme IIB subunit of PTS system from Escherichia coli K12. Northeast Structural Genomics Consortium target ER315/Ontario Center for Structural Proteomics target ec0544 | | Descriptor: | Fructose-like phosphotransferase enzyme IIB component 1 | | Authors: | Wu, B, Yee, A, Gutmanas, A, Semest, A, Montelione, G.T, Arrowsmith, C.H, Northeast Structural Genomics Consortium (NESG), Ontario Centre for Structural Proteomics (OCSP) | | Deposit date: | 2010-06-07 | | Release date: | 2010-06-16 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structure of Enzyme IIB subunit of PTS system from Escherichia coli K12, Northeast Structural Genomics Consortium target ER315/Ontario Center for Structural Proteomics target ec0544
To be Published
|
|
2K4X
 
 | | Solution structure of 30S ribosomal protein S27A from Thermoplasma acidophilum | | Descriptor: | 30S ribosomal protein S27ae, ZINC ION | | Authors: | Wu, B, Yee, A, Fares, C, Lemak, A, Semest, A, Arrowsmith, C, Montelione, G.T, Northeast Structural Genomics Consortium (NESG), Ontario Centre for Structural Proteomics (OCSP) | | Deposit date: | 2008-06-20 | | Release date: | 2008-08-19 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Solution structure of 30S ribosomal protein S27A from Thermoplasma acidophilum/Northeast Structural Genomics Consortium Target TaT88/Ontario Center for Structural Proteomics target ta1093
To be Published
|
|
2KKX
 
 | | Solution Structure of C-terminal domain of reduced NleG2-3 (residues 90-191) from Pathogenic E. coli O157:H7. Northeast Structural Genomics Consortium and Midwest Center for Structural Genomics target ET109A | | Descriptor: | Uncharacterized protein ECs2156 | | Authors: | Wu, B, Yee, A, Fares, C, Lemak, A, Semest, A, Claude, M, Singer, A, Edwards, A, Savchenko, A, Montelione, G.T, Joachimiak, A, Arrowsmith, C.H, Northeast Structural Genomics Consortium (NESG), Ontario Centre for Structural Proteomics (OCSP), Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-06-29 | | Release date: | 2009-08-25 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | NleG Type 3 effectors from enterohaemorrhagic Escherichia coli are U-Box E3 ubiquitin ligases.
Plos Pathog., 6, 2010
|
|
2L2D
 
 | | Solution NMR Structure of human UBA-like domain of OTUD7A_11_83, NESG target HT6304A/OCSP target OTUD7A_11_83/SGC-Toronto | | Descriptor: | OTU domain-containing protein 7A | | Authors: | Wu, B, Yee, A, Lemak, A, Gutmanas, A, Houliston, S, Semesi, A, Dhe-Paganon, S, Montelione, G.T, Arrowsmith, C.H, Northeast Structural Genomics Consortium (NESG), Ontario Centre for Structural Proteomics (OCSP), Structural Genomics Consortium (SGC) | | Deposit date: | 2010-08-17 | | Release date: | 2010-09-01 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The amino-terminal UBA domain of OTUD7A
To be Published
|
|
3LJA
 
 | |
3LEL
 
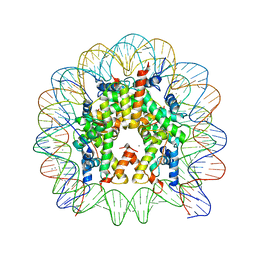 | | Structural Insight into the Sequence-Dependence of Nucleosome Positioning | | Descriptor: | 147-MER DNA, Histone H2A, Histone H2B 1.1, ... | | Authors: | Wu, B, Vasudevan, D, Davey, C.A. | | Deposit date: | 2010-01-15 | | Release date: | 2010-05-19 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Structural insight into the sequence dependence of nucleosome positioning
Structure, 18, 2010
|
|
1SNJ
 
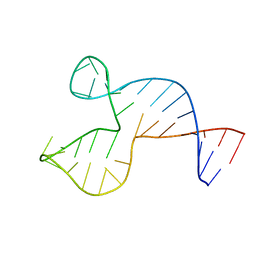 | | Solution structure of the DNA three-way junction with the A/C-stacked conformation | | Descriptor: | 36-MER | | Authors: | Wu, B, Girard, F, van Buuren, B, Schleucher, J, Tessari, M, Wijmenga, S. | | Deposit date: | 2004-03-11 | | Release date: | 2005-04-05 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Global structure of a DNA three-way junction by solution NMR: towards prediction of 3H fold.
Nucleic Acids Res., 32, 2004
|
|
2AKK
 
 | | Solution structure of phnA-like protein rp4479 from Rhodopseudomonas palustris | | Descriptor: | phnA-like protein | | Authors: | Wu, B, Yee, A, Ramelot, T.A, Semesi, A, Lemak, A, Kennedy, M, Edward, A, Arrowsmith, C.H, Northeast Structural Genomics Consortium (NESG), Ontario Centre for Structural Proteomics (OCSP) | | Deposit date: | 2005-08-03 | | Release date: | 2006-08-22 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Solution structure of phnA-like protein rp4479 from Rhodopseudomonas palustris
To be Published
|
|
2AYJ
 
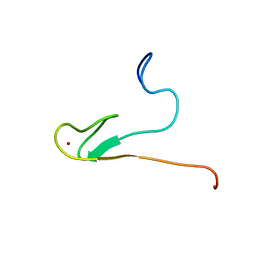 | | Solution structure of 50S ribosomal protein L40e from Sulfolobus solfataricus | | Descriptor: | 50S ribosomal protein L40e, ZINC ION | | Authors: | Wu, B, Yee, A, Lukin, J, Lemak, A, Semesi, A, Ramelot, T, Kennedy, M, Edward, A, Arrowsmith, C.H, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2005-09-07 | | Release date: | 2006-08-22 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of ribosomal protein L40E, a unique C4 zinc finger protein encoded by archaeon Sulfolobus solfataricus
Protein Sci., 17, 2008
|
|
2F1Q
 
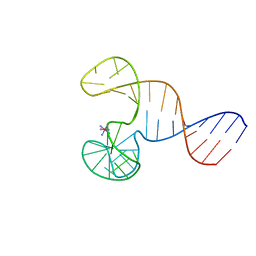 | |
1P5Q
 
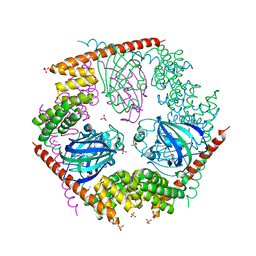 | | Crystal Structure of FKBP52 C-terminal Domain | | Descriptor: | FK506-binding protein 4, SULFATE ION | | Authors: | Wu, B, Li, P, Lou, Z, Shu, C, Ding, Y, Shen, B, Rao, Z. | | Deposit date: | 2003-04-28 | | Release date: | 2004-06-22 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | 3D structure of human FK506-binding protein 52: Implications for the assembly of the glucocorticoid receptor/Hsp90/immunophilin heterocomplex
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
1Q1C
 
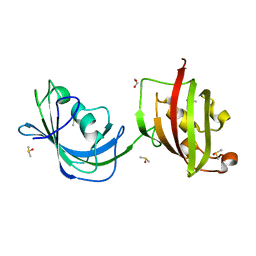 | | Crystal structure of N(1-260) of human FKBP52 | | Descriptor: | 1,2-ETHANEDIOL, DIMETHYL SULFOXIDE, FK506-binding protein 4 | | Authors: | Wu, B, Li, P, Lou, Z, Ding, Y, Shu, C, Shen, B, Rao, Z. | | Deposit date: | 2003-07-18 | | Release date: | 2004-06-22 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | 3D structure of human FK506-binding protein 52: Implications for the assembly of the glucocorticoid receptor/Hsp90/immunophilin heterocomplex
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
1D2B
 
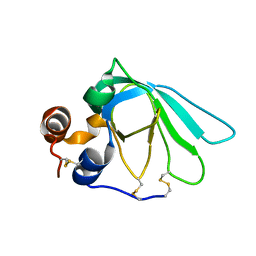 | | THE MMP-INHIBITORY, N-TERMINAL DOMAIN OF HUMAN TISSUE INHIBITOR OF METALLOPROTEINASES-1 (N-TIMP-1), SOLUTION NMR, 29 STRUCTURES | | Descriptor: | Metalloproteinase inhibitor 1 | | Authors: | Wu, B, Arumugam, S, Semenchenko, V, Brew, K, Van Doren, S.R. | | Deposit date: | 1999-09-22 | | Release date: | 1999-12-22 | | Last modified: | 2022-03-23 | | Method: | SOLUTION NMR | | Cite: | NMR structure of tissue inhibitor of metalloproteinases-1 implicates localized induced fit in recognition of matrix metalloproteinases.
J.Mol.Biol., 295, 2000
|
|
2YII
 
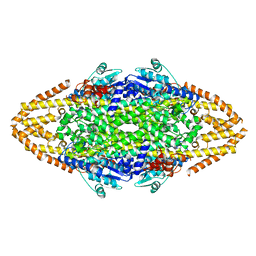 | | Manipulating the regioselectivity of phenylalanine aminomutase: new insights into the reaction mechanism of MIO-dependent enzymes from structure-guided directed evolution | | Descriptor: | BETA-MERCAPTOETHANOL, FORMIC ACID, GLYCEROL, ... | | Authors: | Wu, B, Szymanski, W, Wybenga, G.G, Heberling, M.M, Bartsch, S, Wildeman, S, Poelarends, G.J, Feringa, B.L, Dijkstra, B.W, Janssen, D.B. | | Deposit date: | 2011-05-13 | | Release date: | 2011-11-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Mechanism-Inspired Engineering of Phenylalanine Aminomutase for Enhanced Beta-Regioselective Asymmetric Amination of Cinnamates.
Angew.Chem.Int.Ed.Engl., 51, 2012
|
|
3B6F
 
 | |
3B6G
 
 | |
2LNW
 
 | | Identification and structural basis for a novel interaction between Vav2 and Arap3 | | Descriptor: | Arf-GAP with Rho-GAP domain, ANK repeat and PH domain-containing protein 3, Guanine nucleotide exchange factor VAV2 | | Authors: | Wu, B, Zhang, J, Wu, J, Shi, Y. | | Deposit date: | 2012-01-05 | | Release date: | 2012-11-21 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Identification and structural basis for a novel interaction between Vav2 and Arap3.
J.Struct.Biol., 180, 2012
|
|
2LNX
 
 | | Solution structure of Vav2 SH2 domain | | Descriptor: | Guanine nucleotide exchange factor VAV2 | | Authors: | Wu, B, Zhang, J, Wu, J, Shi, Y. | | Deposit date: | 2012-01-05 | | Release date: | 2012-11-21 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Identification and structural basis for a novel interaction between Vav2 and Arap3.
J.Struct.Biol., 180, 2012
|
|
5AC3
 
 | | Crystal structure of PAM12A | | Descriptor: | ACETIC ACID, CADMIUM ION, PEPTIDE AMIDASE | | Authors: | Wu, B, Wijma, H.J, Song, L, Rozeboom, H.J, Poloni, C, Tian, Y, Arif, M.I, Nuijens, T, Quadflieg, P.J.L.M, Szymanski, W, Feringa, B.L, Janssen, D.B. | | Deposit date: | 2015-08-11 | | Release date: | 2016-07-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Versatile Peptide C-Terminal Functionalization Via a Computationally Peptide Amidase
Acs Catalysis, 2016
|
|
1RQ6
 
 | | Solution structure of ribosomal protein S17E from Methanobacterium Thermoautotrophicum, Northeast Structural Genomics Consortium Target TT802 / Ontario Center for Structural Proteomics Target Mth0803 | | Descriptor: | 30S ribosomal protein S17e | | Authors: | Wu, B, Yee, A, Huang, Y.J, Ramelot, T.A, Semesi, A, Jung, J.W, Edward, A, Lee, W, Kennedy, M.A, Montelione, G.T, Arrowsmith, C.H, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2003-12-04 | | Release date: | 2004-12-14 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The solution structure of ribosomal protein S17E from Methanobacterium thermoautotrophicum: a structural homolog of the FF domain.
Protein Sci., 17, 2008
|
|
1NEI
 
 | | Solution NMR Structure of Protein yoaG from Escherichia coli. Ontario Centre for Structural Proteomics Target EC0264_1_60; Northeast Structural Genomics Consortium Target ET94. | | Descriptor: | hypothetical protein yoaG | | Authors: | Wu, B, Pineda-Lucena, A, Yee, A, Cort, J, Kennedy, M.A, Edwards, A.M, Arrowsmith, C.H, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2002-12-11 | | Release date: | 2004-04-13 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of hypothetical protein dimer encoded by the Yoag gene from Escherichia coli
To be published
|
|
