1DMW
 
 | | CRYSTAL STRUCTURE OF DOUBLE TRUNCATED HUMAN PHENYLALANINE HYDROXYLASE WITH BOUND 7,8-DIHYDRO-L-BIOPTERIN | | Descriptor: | 7,8-DIHYDROBIOPTERIN, FE (III) ION, PHENYLALANINE HYDROXYLASE | | Authors: | Erlandsen, H, Stevens, R.C, Flatmark, T. | | Deposit date: | 1999-12-15 | | Release date: | 2000-03-24 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure and site-specific mutagenesis of pterin-bound human phenylalanine hydroxylase.
Biochemistry, 39, 2000
|
|
5PAH
 
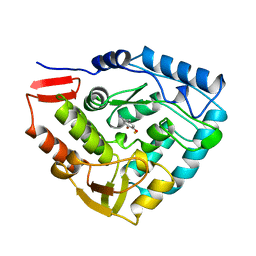 | |
3PAH
 
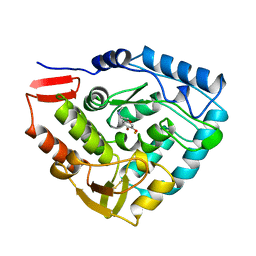 | | HUMAN PHENYLALANINE HYDROXYLASE CATALYTIC DOMAIN DIMER WITH BOUND ADRENALINE INHIBITOR | | Descriptor: | 4-[(1S)-1-hydroxy-2-(methylamino)ethyl]benzene-1,2-diol, FE (III) ION, PHENYLALANINE HYDROXYLASE | | Authors: | Erlandsen, H, Flatmark, T, Stevens, R.C. | | Deposit date: | 1998-08-20 | | Release date: | 1999-04-27 | | Last modified: | 2013-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystallographic analysis of the human phenylalanine hydroxylase catalytic domain with bound catechol inhibitors at 2.0 A resolution.
Biochemistry, 37, 1998
|
|
1TG2
 
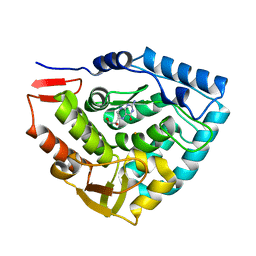 | | Crystal structure of phenylalanine hydroxylase A313T mutant with 7,8-dihydrobiopterin bound | | Descriptor: | 2-AMINO-6-(1,2-DIHYDROXY-PROPYL)-7,8-DIHYDRO-6H-PTERIDIN-4-ONE, FE (III) ION, Phenylalanine-4-hydroxylase | | Authors: | Erlandsen, H, Pey, A.L, Gamez, A, Perez, B, Desviat, L.R, Aguado, C, Koch, R, Surendran, S, Tyring, S, Matalon, R, Scriver, C.R, Ugarte, M, Martinez, A, Stevens, R.C. | | Deposit date: | 2004-05-28 | | Release date: | 2004-11-30 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Correction of kinetic and stability defects by tetrahydrobiopterin in phenylketonuria patients with certain phenylalanine hydroxylase mutations.
Proc.Natl.Acad.Sci.Usa, 101, 2004
|
|
1TDW
 
 | | Crystal structure of double truncated human phenylalanine hydroxylase BH4-responsive PKU mutant A313T. | | Descriptor: | FE (III) ION, Phenylalanine-4-hydroxylase | | Authors: | Erlandsen, H, Pey, A.L, Gamez, A, Perez, B, Desviat, L.R, Aguado, C, Koch, R, Surendran, S, Tyring, S, Matalon, R, Scriver, C.R, Ugarte, M, Martinez, A, Stevens, R.C. | | Deposit date: | 2004-05-24 | | Release date: | 2004-11-30 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Correction of kinetic and stability defects by tetrahydrobiopterin in phenylketonuria patients with certain phenylalanine hydroxylase mutations.
Proc.Natl.Acad.Sci.Usa, 101, 2004
|
|
4PAH
 
 | |
1LTU
 
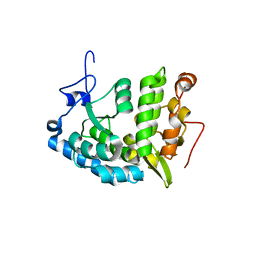 | | CRYSTAL STRUCTURE OF CHROMOBACTERIUM VIOLACEUM, APO (NO IRON BOUND) STRUCTURE | | Descriptor: | PHENYLALANINE-4-HYDROXYLASE | | Authors: | Erlandsen, H, Kim, J.Y, Patch, M.G, Han, A, Volner, A, Abu-Omar, M.M, Stevens, R.C. | | Deposit date: | 2002-05-20 | | Release date: | 2002-07-17 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Structural comparison of bacterial and human iron-dependent phenylalanine hydroxylases: similar fold, different stability and reaction rates.
J.Mol.Biol., 320, 2002
|
|
1LTZ
 
 | | CRYSTAL STRUCTURE OF CHROMOBACTERIUM VIOLACEUM PHENYLALANINE HYDROXYLASE, STRUCTURE HAS BOUND IRON (III) AND OXIDIZED COFACTOR 7,8-DIHYDROBIOPTERIN | | Descriptor: | 7,8-DIHYDROBIOPTERIN, CHLORIDE ION, FE (III) ION, ... | | Authors: | Erlandsen, H, Kim, J.Y, Patch, M.G, Han, A, Volner, A, Abu-Omar, M.M, Stevens, R.C. | | Deposit date: | 2002-05-21 | | Release date: | 2002-07-17 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural comparison of bacterial and human iron-dependent phenylalanine hydroxylases: similar fold, different stability and reaction rates.
J.Mol.Biol., 320, 2002
|
|
1LTV
 
 | | CRYSTAL STRUCTURE OF CHROMOBACTERIUM VIOLACEUM PHENYLALANINE HYDROXYLASE, STRUCTURE WITH BOUND OXIDIZED Fe(III) | | Descriptor: | FE (III) ION, PHENYLALANINE-4-HYDROXYLASE | | Authors: | Erlandsen, H, Kim, J.Y, Patch, M.G, Han, A, Volner, A, Abu-Omar, M.M, Stevens, R.C. | | Deposit date: | 2002-05-20 | | Release date: | 2002-07-17 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural comparison of bacterial and human iron-dependent phenylalanine hydroxylases: similar fold, different stability and reaction rates.
J.Mol.Biol., 320, 2002
|
|
6PAH
 
 | | HUMAN PHENYLALANINE HYDROXYLASE CATALYTIC DOMAIN DIMER WITH BOUND L-DOPA (3,4-DIHYDROXYPHENYLALANINE) INHIBITOR | | Descriptor: | 3,4-DIHYDROXYPHENYLALANINE, FE (III) ION, PHENYLALANINE 4-MONOOXYGENASE | | Authors: | Erlandsen, H, Flatmark, T, Stevens, R.C. | | Deposit date: | 1998-08-20 | | Release date: | 1999-04-27 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystallographic analysis of the human phenylalanine hydroxylase catalytic domain with bound catechol inhibitors at 2.0 A resolution.
Biochemistry, 37, 1998
|
|
1PAH
 
 | |
2FRX
 
 | | Crystal structure of YebU, a m5C RNA methyltransferase from E.coli | | Descriptor: | Hypothetical protein yebU | | Authors: | Erlandsen, H, Nordlund, P, Hallberg, B.M, Johnson, K.A, Ericsson, U.B. | | Deposit date: | 2006-01-20 | | Release date: | 2006-08-29 | | Last modified: | 2018-05-23 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The structure of the RNA m5C methyltransferase YebU from Escherichia coli reveals a C-terminal RNA-recruiting PUA domain
J.Mol.Biol., 360, 2006
|
|
7MYM
 
 | | Crystal structure of Escherichia coli dihydrofolate reductase in complex with TRIMETHOPRIM and NADPH | | Descriptor: | ARGININE, Dihydrofolate reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Erlandsen, H, Wright, D, Krucinska, J. | | Deposit date: | 2021-05-21 | | Release date: | 2022-06-01 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.04 Å) | | Cite: | Structure-guided functional studies of plasmid-encoded dihydrofolate reductases reveal a common mechanism of trimethoprim resistance in Gram-negative pathogens.
Commun Biol, 5, 2022
|
|
7MYL
 
 | |
6BFZ
 
 | | Crystal structure of enolase from E. coli with a mixture of apo form, substrate, and product form | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-PHOSPHOGLYCERIC ACID, Enolase, ... | | Authors: | Erlandsen, H, Wright, D, Krucinska, J. | | Deposit date: | 2017-10-27 | | Release date: | 2018-10-31 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Structural and Functional Studies of Bacterial Enolase, a Potential Target against Gram-Negative Pathogens.
Biochemistry, 58, 2019
|
|
6BFY
 
 | | Crystal structure of enolase from Escherichia coli with bound 2-phosphoglycerate substrate | | Descriptor: | 2-PHOSPHOGLYCERIC ACID, Enolase, GLYCEROL, ... | | Authors: | Erlandsen, H, Wright, D, Krucinska, J. | | Deposit date: | 2017-10-27 | | Release date: | 2018-10-31 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Structural and Functional Studies of Bacterial Enolase, a Potential Target against Gram-Negative Pathogens.
Biochemistry, 58, 2019
|
|
6NPF
 
 | | Structure of E.coli enolase in complex with an analog of the natural product SF-2312 metabolite. | | Descriptor: | Enolase, GLYCEROL, L(+)-TARTARIC ACID, ... | | Authors: | Erlandsen, H, Krucinska, J, Lombardo, M, Wright, D. | | Deposit date: | 2019-01-17 | | Release date: | 2019-11-27 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | Functional and structural basis of E. coli enolase inhibition by SF2312: a mimic of the carbanion intermediate.
Sci Rep, 9, 2019
|
|
6D3L
 
 | | Crystal structure of unphosphorylated human PKR | | Descriptor: | Interferon-induced, double-stranded RNA-activated protein kinase | | Authors: | Erlandsen, H, Mayo, C, Robinson, V.L, Cole, J.L. | | Deposit date: | 2018-04-16 | | Release date: | 2019-07-10 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural Basis of Protein Kinase R Autophosphorylation.
Biochemistry, 58, 2019
|
|
6D3Q
 
 | | Crystal structure of Escherichia coli enolase complexed with a natural inhibitor SF2312. | | Descriptor: | Enolase, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Erlandsen, H, Krucinska, J, Hazeen, A, Wright, D. | | Deposit date: | 2018-04-16 | | Release date: | 2019-11-27 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Functional and structural basis of E. coli enolase inhibition by SF2312: a mimic of the carbanion intermediate.
Sci Rep, 9, 2019
|
|
6D3K
 
 | | Crystal structure of unphosphorylated human PKR kinase domain in complex with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Interferon-induced, double-stranded RNA-activated protein kinase, ... | | Authors: | Erlandsen, H, Mayo, C.B, Robinson, V.L, Cole, J.L. | | Deposit date: | 2018-04-16 | | Release date: | 2019-07-10 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural Basis of Protein Kinase R Autophosphorylation.
Biochemistry, 58, 2019
|
|
3FHD
 
 | | Crystal structure of the Shutoff and Exonuclease Protein from Kaposis Sarcoma Associated Herpesvirus | | Descriptor: | MAGNESIUM ION, ORF 37, SULFATE ION | | Authors: | Dahlroth, S.L, Gurmu, D, Schmitzberger, F, Haas, J, Erlandsen, H, Nordlund, P. | | Deposit date: | 2008-12-09 | | Release date: | 2009-11-24 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of the shutoff and exonuclease protein from the oncogenic Kaposi's sarcoma-associated herpesvirus
Febs J., 276, 2009
|
|
2VEC
 
 | | The crystal structure of the protein YhaK from Escherichia coli | | Descriptor: | CHLORIDE ION, PIRIN-LIKE PROTEIN YHAK | | Authors: | Gurmu, D, Lu, J, Johnson, K.A, Nordlund, P, Holmgren, A, Erlandsen, H. | | Deposit date: | 2007-10-18 | | Release date: | 2008-07-01 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The Crystal Structure of the Protein Yhak from Escherichia Coli Reveals a New Subclass of Redox Sensitive Enterobacterial Bicupins.
Proteins, 74, 2008
|
|
2PAH
 
 | |
3QKW
 
 | | Structure of Streptococcus parasangunini Gtf3 glycosyltransferase | | Descriptor: | Nucleotide sugar synthetase-like protein, URIDINE-5'-DIPHOSPHATE | | Authors: | Zhu, F, Erlandsen, H, Huang, Y, Ding, L, Zhou, M, Liang, X, Ma, J.-B, Wu, H. | | Deposit date: | 2011-02-01 | | Release date: | 2011-06-08 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.287 Å) | | Cite: | Structural and Functional Analysis of a New Subfamily of Glycosyltransferases Required for Glycosylation of Serine-rich Streptococcal Adhesins.
J.Biol.Chem., 286, 2011
|
|
7NAE
 
 | | Crystal structure of Escherichia coli dihydrofolate reductase in complex with TRIMETHOPRIM | | Descriptor: | Dihydrofolate reductase, SULFATE ION, TRIMETHOPRIM | | Authors: | Estrada, A, Wright, D, Krucinska, J, Erlandsen, H. | | Deposit date: | 2021-06-21 | | Release date: | 2022-06-29 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structure-guided functional studies of plasmid-encoded dihydrofolate reductases reveal a common mechanism of trimethoprim resistance in Gram-negative pathogens.
Commun Biol, 5, 2022
|
|
