4AY9
 
 | | Structure of follicle-stimulating hormone in complex with the entire ectodomain of its receptor | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, FOLLICLE-STIMULATING HORMONE RECEPTOR, FOLLITROPIN SUBUNIT BETA, ... | | Authors: | Jiang, X, Liu, H, Chen, X, He, X. | | Deposit date: | 2012-06-19 | | Release date: | 2012-08-08 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of Follicle-Stimulating Hormone in Complex with the Entire Ectodomain of its Receptor.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
1GKO
 
 | | An Engineered Transthyretin Monomer that is Non-amyloidogenic - Unless Partially Denatured | | Descriptor: | TRANSTHYRETIN | | Authors: | Jiang, X, Smith, C.S, Petrassi, H.M, Hammarstrom, P, White, J.T, Sacchettini, J.C, Kelly, J.W. | | Deposit date: | 2001-08-17 | | Release date: | 2001-11-28 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | An Engineered Transthyretin Monomer that is Nonamyloidogenic, Unless It is Partially Denatured
Biochemistry, 40, 2001
|
|
6N3I
 
 | |
1SCF
 
 | | HUMAN RECOMBINANT STEM CELL FACTOR | | Descriptor: | CALCIUM ION, PENTAETHYLENE GLYCOL, STEM CELL FACTOR | | Authors: | Jiang, X, Gurel, O, Langley, K.E, Hendrickson, W.A. | | Deposit date: | 1998-06-04 | | Release date: | 2000-07-07 | | Last modified: | 2022-12-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of the active core of human stem cell factor and analysis of binding to its receptor kit.
EMBO J., 19, 2000
|
|
3WZF
 
 | | Crystal structure of human cytoplasmic aspartate aminotransferase | | Descriptor: | Aspartate aminotransferase, cytoplasmic | | Authors: | Jiang, X, Chang, H, Zhou, Y, Chen, L, Yang, Q. | | Deposit date: | 2014-09-24 | | Release date: | 2015-10-07 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.991 Å) | | Cite: | Recombinant expression, purification and Preliminary crystallographic studies of human cytoplasmic aspartate aminotransferase
To be Published
|
|
5AX8
 
 | | Recombinant expression, purification and preliminary crystallographic studies of the mature form of human mitochondrial aspartate aminotransferase | | Descriptor: | Aspartate aminotransferase, mitochondrial | | Authors: | Jiang, X, Wang, J, Chang, H, Zhou, Y. | | Deposit date: | 2015-07-20 | | Release date: | 2016-03-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.989 Å) | | Cite: | Recombinant expression, purification and crystallographic studies of the mature form of human mitochondrial aspartate aminotransferase
Biosci Trends, 10, 2016
|
|
4G9Z
 
 | | Lassa nucleoprotein with dsRNA reveals novel mechanism for immune suppression | | Descriptor: | Nucleoprotein, RNA (5'-R(P*GP*CP*CP*C)-3'), RNA (5'-R(P*GP*GP*GP*C)-3'), ... | | Authors: | Jiang, X, Huang, Q, Wang, W, Dong, H, Ly, H, Liang, Y, Dong, C, Scottish Structural Proteomics Facility (SSPF) | | Deposit date: | 2012-07-24 | | Release date: | 2013-04-03 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Structures of Arenaviral Nucleoproteins with Triphosphate dsRNA Reveal a Unique Mechanism of Immune Suppression.
J.Biol.Chem., 288, 2013
|
|
4GV6
 
 | | Structures of Lassa and Tacaribe viral nucleoproteins with or without 5 triphosphate dsRNA substrate reveal a unique 3 -5 exoribonuclease mechanism to suppress type I interferon production | | Descriptor: | MANGANESE (II) ION, Nucleoprotein, RNA (5'-R(*(GTP)P*GP*GP*C)-3'), ... | | Authors: | Jiang, X, Huang, Q, Wang, W, Dong, H, Ly, H, Liang, Y, Dong, C. | | Deposit date: | 2012-08-30 | | Release date: | 2013-05-01 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Structures of Arenaviral Nucleoproteins with Triphosphate dsRNA Reveal a Unique Mechanism of Immune Suppression.
J.Biol.Chem., 288, 2013
|
|
4GVE
 
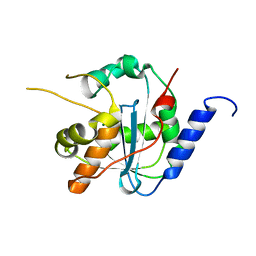 | | Tacaribe nucleoprotein structure | | Descriptor: | MAGNESIUM ION, Nucleoprotein, ZINC ION | | Authors: | Jiang, X, Huang, Q, Wang, W, Dong, H, Ly, H, Liang, Y, Dong, C. | | Deposit date: | 2012-08-30 | | Release date: | 2013-05-01 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Structures of Arenaviral Nucleoproteins with Triphosphate dsRNA Reveal a Unique Mechanism of Immune Suppression.
J.Biol.Chem., 288, 2013
|
|
4GV9
 
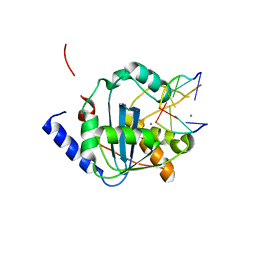 | | Lassa nucleoprotein C-terminal domain in complex with triphosphated dsRNA soaking for 5 min | | Descriptor: | MANGANESE (II) ION, Nucleoprotein, RNA (5'-R(*(GTP)P*GP*GP*C)-3'), ... | | Authors: | Jiang, X, Huang, Q, Wang, W, Dong, H, Ly, H, Liang, Y, Dong, C. | | Deposit date: | 2012-08-30 | | Release date: | 2013-05-01 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | Structures of Arenaviral Nucleoproteins with Triphosphate dsRNA Reveal a Unique Mechanism of Immune Suppression.
J.Biol.Chem., 288, 2013
|
|
4MQW
 
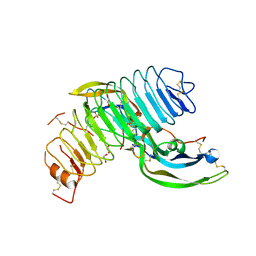 | | Structure of follicle-stimulating hormone in complex with the entire ectodomain of its receptor (P31) | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, Follicle-stimulating hormone receptor, ... | | Authors: | Jiang, X, Liu, H, Chen, X, He, X. | | Deposit date: | 2013-09-16 | | Release date: | 2014-04-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Evidence for Follicle-stimulating Hormone Receptor as a Functional Trimer.
J.Biol.Chem., 289, 2014
|
|
4GV3
 
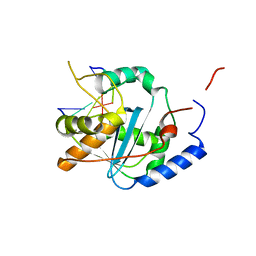 | | Structures of Lassa and Tacaribe viral nucleoproteins with or without 5 triphosphate dsRNA substrate reveal a unique 3 -5 exoribonuclease mechanism to suppress type I interferon production | | Descriptor: | MANGANESE (II) ION, Nucleoprotein, RNA (5'-R(*(GTP)P*GP*GP*C)-3'), ... | | Authors: | Jiang, X, Huang, Q, Wang, W, Dong, H, Ly, H, Liang, Y, Dong, C. | | Deposit date: | 2012-08-30 | | Release date: | 2013-05-01 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Structures of Arenaviral Nucleoproteins with Triphosphate dsRNA Reveal a Unique Mechanism of Immune Suppression.
J.Biol.Chem., 288, 2013
|
|
5GXB
 
 | | crystal structure of a LacY/Nanobody complex | | Descriptor: | Lactose permease, nanobody | | Authors: | Jiang, X, Wu, J.P, Yan, N, Kaback, H.R. | | Deposit date: | 2016-09-16 | | Release date: | 2016-10-26 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Crystal structure of a LacY-nanobody complex in a periplasmic-open conformation.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
6IZO
 
 | | Crystal structure of DNA polymerase sliding clamp from Caulobacter crescentus | | Descriptor: | 1,2-ETHANEDIOL, Beta sliding clamp, DI(HYDROXYETHYL)ETHER | | Authors: | Jiang, X, Zhang, L, Teng, M, Li, X. | | Deposit date: | 2018-12-20 | | Release date: | 2019-11-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Caulobacter crescentus beta sliding clamp employs a noncanonical regulatory model of DNA replication.
Febs J., 287, 2020
|
|
7CLA
 
 | |
6JIR
 
 | | Crystal structure of C. crescentus beta sliding clamp with PEG bound to putative beta-motif tethering region | | Descriptor: | 1,2-ETHANEDIOL, Beta sliding clamp, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Jiang, X, Teng, M, Li, X. | | Deposit date: | 2019-02-23 | | Release date: | 2019-11-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Caulobacter crescentus beta sliding clamp employs a noncanonical regulatory model of DNA replication.
Febs J., 287, 2020
|
|
6M2L
 
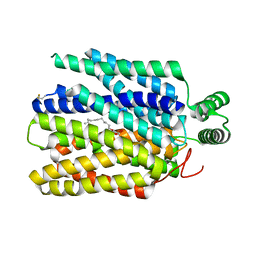 | | Crystal structure of Plasmodium falciparum hexose transporter PfHT1 bound with C3361 | | Descriptor: | (2S,3R,4S,5R,6R)-6-(hydroxymethyl)-4-undec-10-enoxy-oxane-2,3,5-triol, Hexose transporter 1 | | Authors: | Jiang, X, Yuan, Y.Y, Zhang, S, Wang, N, Yan, C.Y, Yan, N. | | Deposit date: | 2020-02-27 | | Release date: | 2020-09-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Structural Basis for Blocking Sugar Uptake into the Malaria Parasite Plasmodium falciparum.
Cell, 183, 2020
|
|
6M20
 
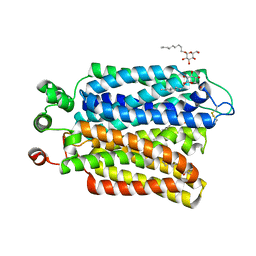 | | Crystal structure of Plasmodium falciparum hexose transporter PfHT1 bound with glucose | | Descriptor: | Hexose transporter 1, beta-D-glucopyranose, nonyl beta-D-glucopyranoside | | Authors: | Jiang, X, Yuan, Y.Y, Zhang, S, Wang, N, Yan, C.Y, Yan, N. | | Deposit date: | 2020-02-26 | | Release date: | 2020-09-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural Basis for Blocking Sugar Uptake into the Malaria Parasite Plasmodium falciparum.
Cell, 183, 2020
|
|
6JMN
 
 | | Crystal structure of Ostrinia furnacalis Chitinase h complexed with compound 2-8-s2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 6-azanyl-11-methyl-2-oxidanylidene-7-[[(2R)-oxolan-2-yl]methyl]-N-(pyridin-3-ylmethyl)-1,9-diaza-7-azoniatricyclo[8.4.0.0^{3,8}]tetradeca-3(8),4,6,9,11,13-hexaene-5-carboxamide, Chitinase | | Authors: | Jiang, X, Yang, Q. | | Deposit date: | 2019-03-12 | | Release date: | 2020-05-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A Series of Compounds Bearing a Dipyrido-Pyrimidine Scaffold Acting as Novel Human and Insect Pest Chitinase Inhibitors.
J.Med.Chem., 63, 2020
|
|
6JJR
 
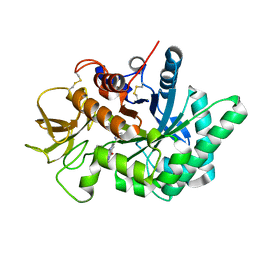 | | Crystal structure of human chitotriosidase-1 (hChit1) catalytic domain in complex with compound 2-8-14 | | Descriptor: | 6-azanyl-2-oxidanylidene-N-[(1S)-1-phenylethyl]-7-(phenylmethyl)-1$l^{4},9-diaza-7-azoniatricyclo[8.4.0.0^{3,8}]tetradeca-1(14),3(8),4,6,10,12-hexaene-5-carboxamide, Chitotriosidase-1 | | Authors: | Jiang, X, Yang, Q. | | Deposit date: | 2019-02-26 | | Release date: | 2020-02-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.834 Å) | | Cite: | A Series of Compounds Bearing a Dipyrido-Pyrimidine Scaffold Acting as Novel Human and Insect Pest Chitinase Inhibitors.
J.Med.Chem., 63, 2020
|
|
6JK9
 
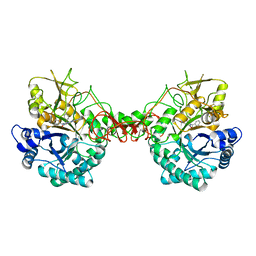 | | Crystal structure of Serratia marcescens Chitinase B complexed with compound 2-8-14 | | Descriptor: | 6-azanyl-2-oxidanylidene-N-[(1S)-1-phenylethyl]-7-(phenylmethyl)-1$l^{4},9-diaza-7-azoniatricyclo[8.4.0.0^{3,8}]tetradeca-1(14),3(8),4,6,10,12-hexaene-5-carboxamide, Chitinase | | Authors: | Jiang, X, Yang, Q. | | Deposit date: | 2019-02-27 | | Release date: | 2020-02-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.312 Å) | | Cite: | A Series of Compounds Bearing a Dipyrido-Pyrimidine Scaffold Acting as Novel Human and Insect Pest Chitinase Inhibitors.
J.Med.Chem., 63, 2020
|
|
7CB1
 
 | | Structure of Serratia marcescens chitinase B complexed with compound 6q | | Descriptor: | 6-azanyl-11-methyl-2-oxidanylidene-N-[(1S)-1-pyridin-3-ylethyl]-1,7,9-triazatricyclo[8.4.0.0^{3,8}]tetradeca-3,5,7,9,11,13-hexaene-5-carboxamide, Chitinase B | | Authors: | Jiang, X, Yuan, P.T. | | Deposit date: | 2020-06-10 | | Release date: | 2021-06-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Discovery of novel Chitinase Inhibitors with a scaffold of dipyridopyrimidine-3-carboxamide via Structure-based Rational Design
To Be Published
|
|
6JK6
 
 | | Crystal structure of human chitotriosidase-1 (hCHIT) catalytic domain in complex with compound 2-8-s2 | | Descriptor: | 6-azanyl-11-methyl-2-oxidanylidene-7-[[(2R)-oxolan-2-yl]methyl]-N-(pyridin-3-ylmethyl)-1,9-diaza-7-azoniatricyclo[8.4.0.0^{3,8}]tetradeca-3(8),4,6,9,11,13-hexaene-5-carboxamide, Chitotriosidase-1 | | Authors: | Jiang, X, Yang, Q. | | Deposit date: | 2019-02-27 | | Release date: | 2020-02-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.571 Å) | | Cite: | A Series of Compounds Bearing a Dipyrido-Pyrimidine Scaffold Acting as Novel Human and Insect Pest Chitinase Inhibitors.
J.Med.Chem., 63, 2020
|
|
4JAJ
 
 | | Crystal Structure of Aurora Kinase A in complex with BENZO[C][1,8]NAPHTHYRIDIN-6(5H)-ONE | | Descriptor: | Aurora kinase A, benzo[c][1,8]naphthyridin-6(5H)-one | | Authors: | Jiang, X, Josephson, K, Huck, B, Goutopoulos, A, Karra, S. | | Deposit date: | 2013-02-18 | | Release date: | 2013-05-22 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | SAR and evaluation of novel 5H-benzo[c][1,8]naphthyridin-6-one analogs as Aurora kinase inhibitors.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
4JAI
 
 | | Crystal Structure of Aurora Kinase A in complex with N-{4-[(6-oxo-5,6-dihydrobenzo[c][1,8]naphthyridin-1-yl)amino]phenyl}benzamide | | Descriptor: | Aurora kinase A, N-{4-[(6-oxo-5,6-dihydrobenzo[c][1,8]naphthyridin-1-yl)amino]phenyl}benzamide | | Authors: | Jiang, X, Josephson, K, Huck, B, Goutopoulos, A, Karra, S. | | Deposit date: | 2013-02-18 | | Release date: | 2013-05-22 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | SAR and evaluation of novel 5H-benzo[c][1,8]naphthyridin-6-one analogs as Aurora kinase inhibitors.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
