2QS6
 
 | | Structure of a Hoogsteen antiparallel duplex with extra-helical thymines | | Descriptor: | DNA (5'-D(*DAP*DTP*DAP*DTP*DAP*DTP*DCP*DT)-3') | | Authors: | Pous, J, Urpi, L, Subirana, J.A, Gouyette, C, Navaza, J, Campos, J.L. | | Deposit date: | 2007-07-30 | | Release date: | 2008-03-25 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.08 Å) | | Cite: | Stabilization by extra-helical thymines of a DNA duplex with Hoogsteen base pairs.
J.Am.Chem.Soc., 130, 2008
|
|
8ACM
 
 | | Crystal structure of WT p38alpha | | Descriptor: | 4-[5-(4-FLUORO-PHENYL)-2-(4-METHANESULFINYL-PHENYL)-3H-IMIDAZOL-4-YL]-PYRIDINE, MAGNESIUM ION, Mitogen-activated protein kinase 14 | | Authors: | Pous, J, Baginski, B, Gonzalez, L, Macias, M.J, Nebreda, A.R. | | Deposit date: | 2022-07-05 | | Release date: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Crystal structure of WT p38alpha
Res Sq
|
|
8ACO
 
 | | Crystal structure of WT p38alpha | | Descriptor: | 4-[5-(4-FLUORO-PHENYL)-2-(4-METHANESULFINYL-PHENYL)-3H-IMIDAZOL-4-YL]-PYRIDINE, MAGNESIUM ION, Mitogen-activated protein kinase 14 | | Authors: | Pous, J, Baginski, B, Gonzalez, L, Macias, M.J, Nebreda, A.R. | | Deposit date: | 2022-07-05 | | Release date: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Crystal structure of WT p38alpha
Res Sq
|
|
1E21
 
 | | Ribonuclease 1 des1-7 Crystal Structure at 1.9A | | Descriptor: | RIBONUCLEASE 1 | | Authors: | Pous, J, Mallorqui-Fernandez, G, Peracaula, R, Terzyan, S.S, Futami, J, Tada, H, Yamada, H, Seno, M, De Llorens, R, Gomis-Ruth, F.X, Coll, M. | | Deposit date: | 2000-05-15 | | Release date: | 2001-05-03 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Three-Dimensional Crystal Structure of Human Rnase 1Dn7 at 1.9A Resolution
Acta Crystallogr.,Sect.D, 57, 2001
|
|
1DZA
 
 | | 3-D structure of a HP-RNase | | Descriptor: | RIBONUCLEASE 1 | | Authors: | Pous, J, Canals, A, Terzyan, S.S, Guasch, A, Benito, A, Ribo, M, Vilanova, M, Coll, M. | | Deposit date: | 2000-02-21 | | Release date: | 2001-02-16 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Three-Dimensional Structure of a Human Pancreatic Ribonuclease Variant, a Step Forward in the Design of Cytotoxic Ribonucleases
J.Mol.Biol., 303, 2000
|
|
8PQN
 
 | | NQO1 bound to RBS-10 | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, NAD(P)H dehydrogenase [quinone] 1, ~{N}-[4-[(3-methylphenyl)carbonylamino]phenyl]-5-nitro-furan-2-carboxamide | | Authors: | Pous, J, Jose-Duran, F, Mayor-Ruiz, C, Riera, A. | | Deposit date: | 2023-07-11 | | Release date: | 2024-01-10 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Discovery and Mechanistic Elucidation of NQO1-Bioactivatable Small Molecules That Overcome Resistance to Degraders.
Angew.Chem.Int.Ed.Engl., 63, 2024
|
|
3EY0
 
 | | A new form of DNA-drug interaction in the minor groove of a coiled coil | | Descriptor: | 1,5-BIS(4-AMIDINOPHENOXY)PENTANE, 5'-D(*DAP*DTP*DAP*DTP*DAP*DTP*DAP*DTP*DAP*DT)-3', MAGNESIUM ION | | Authors: | Pous, J, Moreno, T, Subirana, J.A, Campos, J.L. | | Deposit date: | 2008-10-17 | | Release date: | 2009-10-27 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | Coiled-coil conformation of a pentamidine-DNA complex
Acta Crystallogr.,Sect.D, 66, 2010
|
|
3NAP
 
 | | Structure of Triatoma Virus (TrV) | | Descriptor: | Capsid protein | | Authors: | Squires, G, Pous, J. | | Deposit date: | 2010-06-02 | | Release date: | 2010-07-28 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The crystallographic structure of Triatoma Virus (TrV) highlights the Dicistroviridae and Picornaviridae family differences
To be Published
|
|
7PVU
 
 | | Crystal structure of p38alpha C162S in complex with CAS2094511-69-8, P 1 21 1 | | Descriptor: | Mitogen-activated protein kinase 14, N-(2-cyclobutyl-1H-1,3-benzodiazol-5-yl)-2-fluorobenzene-1-sulfonamide | | Authors: | Baginski, B, Pous, J, Gonzalez, L, Macias, M.J, Nebreda, A.R. | | Deposit date: | 2021-10-05 | | Release date: | 2023-05-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.154 Å) | | Cite: | Characterization of p38 alpha autophosphorylation inhibitors that target the non-canonical activation pathway.
Nat Commun, 14, 2023
|
|
2GSY
 
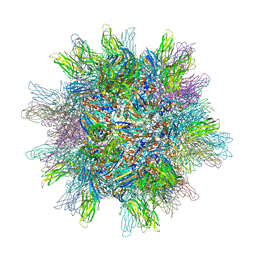 | | The 2.6A structure of Infectious Bursal Virus Derived T=1 Particles | | Descriptor: | CALCIUM ION, polyprotein | | Authors: | Garriga, D, Querol-Audi, J, Abaitua, F, Saugar, I, Pous, J, Verdaguer, N, Caston, J.R, Rodriguez, J.F. | | Deposit date: | 2006-04-27 | | Release date: | 2006-07-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The 2.6-angstrom structure of infectious bursal disease virus-derived t=1 particles reveals new stabilizing elements of the virus capsid.
J.Virol., 80, 2006
|
|
1WCD
 
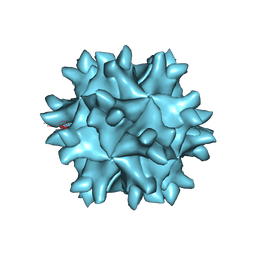 | | Crystal structure of IBDV T1 virus-like particle reveals a missing link in icosahedral viruses evolution | | Descriptor: | MAJOR STRUCTURAL PROTEIN VP2 | | Authors: | Coulibaly, F, Chevalier, C, Gutsche, I, Pous, J, Bressanelli, S, Navaza, J, Delmas, B, Rey, F.A. | | Deposit date: | 2004-11-12 | | Release date: | 2005-04-05 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The Birnavirus Crystal Structure Reveals Structural Relationships Among Icosahedral Viruses.
Cell(Cambridge,Mass.), 120, 2005
|
|
1WCE
 
 | | Crystal structure of the T13 IBDV viral particle reveals a missing link in icosahedral viruses evolution | | Descriptor: | MAJOR STRUCTURAL PROTEIN VP2 | | Authors: | Coulibaly, F, Chevalier, C, Gutsche, I, Pous, J, Bressanelli, S, Navaza, J, Delmas, B, Rey, F.A. | | Deposit date: | 2004-11-12 | | Release date: | 2005-04-06 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (7 Å) | | Cite: | The Birnavirus Crystal Structure Reveals Structural Relationships Among Icosahedral Viruses.
Cell(Cambridge,Mass.), 120, 2005
|
|
3UXW
 
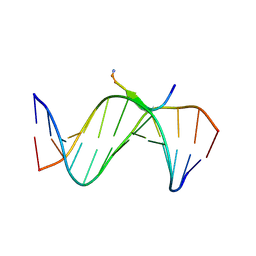 | | Crystal Structures of an A-T-hook/DNA complex | | Descriptor: | A-T hook peptide, dodecamer DNA | | Authors: | Fonfria-Subiros, E, Acosta-Reyes, F.J, Saperas, N, Pous, J, Subirana, J.A, Campos, J.L. | | Deposit date: | 2011-12-05 | | Release date: | 2012-05-23 | | Last modified: | 2013-03-27 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Crystal structure of a complex of DNA with one AT-hook of HMGA1.
Plos One, 7, 2012
|
|
4HL8
 
 | | Re-refinement of the vault ribonucleoprotein particle | | Descriptor: | Major vault protein | | Authors: | Casanas, A, Querol-Audi, J, Guerra, P, Pous, J, Tanaka, H, Tsukihara, T, Verdaguer, V, Fita, I. | | Deposit date: | 2012-10-16 | | Release date: | 2013-06-05 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | New features of vault architecture and dynamics revealed by novel refinement using the deformable elastic network approach.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
3DPR
 
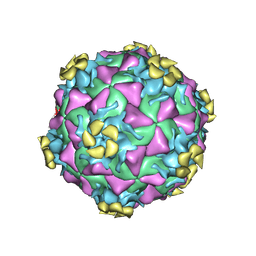 | | Human rhinovirus 2 bound to a concatamer of the VLDL receptor module V3 | | Descriptor: | CALCIUM ION, LAURIC ACID, LDL-receptor class A 3, ... | | Authors: | Querol-Audi, J, Pous, J, Fita, I, Verdaguer, N. | | Deposit date: | 2008-07-09 | | Release date: | 2009-04-07 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Minor group human rhinovirus-receptor interactions: geometry of multimodular attachment and basis of recognition
Febs Lett., 583, 2009
|
|
6TJP
 
 | | Crystal structure of T7 bacteriophage portal protein, 13mer, closed valve - P212121 | | Descriptor: | Portal protein | | Authors: | Fabrega-Ferrer, M, Cuervo, A, Fernandez, F.J, Machon, C, Perez-Luque, R, Pous, J, Vega, M.C, Carrascosa, J.L, Coll, M. | | Deposit date: | 2019-11-26 | | Release date: | 2020-12-16 | | Last modified: | 2021-06-30 | | Method: | X-RAY DIFFRACTION (3.74 Å) | | Cite: | Using a partial atomic model from medium-resolution cryo-EM to solve a large crystal structure.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
1H8X
 
 | | Domain-swapped Dimer of a Human Pancreatic Ribonuclease Variant | | Descriptor: | RIBONUCLEASE 1 | | Authors: | Canals, A, Pous, J, Guasch, A, Benito, A, Ribo, M, Vilanova, M, Coll, M. | | Deposit date: | 2001-02-16 | | Release date: | 2002-02-14 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Structure of an Engineered Domain-Swapped Ribonuclease Dimer and its Implications for the Evolution of Proteins Toward Oligomerization
Structure, 9, 2001
|
|
4AY3
 
 | | Crystal structure of Bacillus anthracis PurE | | Descriptor: | ACETATE ION, N5-CARBOXYAMINOIMIDAZOLE RIBONUCLEOTIDE MUTASE | | Authors: | Oliete, R, Pous, J, Rodriguez-Puente, S, Abad-Zapatero, C, Guasch, A. | | Deposit date: | 2012-06-18 | | Release date: | 2013-01-30 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Elastic and Inelastic Diffraction Changes Upon Variation of the Relative Humidity Environment of Pure Crystals
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4AY4
 
 | | crystal structure of Bacillus anthracis PurE | | Descriptor: | ACETATE ION, N5-CARBOXYAMINOIMIDAZOLE RIBONUCLEOTIDE MUTASE | | Authors: | Oliete, R, Pous, J, Rodriguez-Puente, S, Abad-Zapatero, C, Guasch, A. | | Deposit date: | 2012-06-18 | | Release date: | 2013-01-30 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Elastic and Inelastic Diffraction Changes Upon Variation of the Relative Humidity Environment of Pure Crystals
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4LT7
 
 | | Crystal structure of the c2a domain of rabphilin-3a in complex with a calcium | | Descriptor: | CALCIUM ION, Rabphilin-3A | | Authors: | Verdaguer, N, Ferrer-Orta, C, Buxaderas, M, Corbalan-Garcia, S, Perez-Sanchez, D, Guerrero-Valero, M, Luengo, G, Pous, J, Guerra, P, Gomez-Fernandez, J.C, Guillen, J. | | Deposit date: | 2013-07-23 | | Release date: | 2013-12-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural insights into the Ca2+ and PI(4,5)P2 binding modes of the C2 domains of rabphilin 3A and synaptotagmin 1.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4B4K
 
 | | Crystal structure of Bacillus anthracis PurE | | Descriptor: | N5-CARBOXYAMINOIMIDAZOLE RIBONUCLEOTIDE MUTASE | | Authors: | Oliete, R, Pous, J, Rodriguez-Puente, S, Abad-Zapatero, C, Guasch, A. | | Deposit date: | 2012-07-31 | | Release date: | 2012-08-08 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Elastic and Inelastic Diffraction Changes Upon Variation of the Relative Humidity Environment of Pure Crystals
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4NS0
 
 | | The C2A domain of Rabphilin 3A in complex with PI(4,5)P2 | | Descriptor: | Rabphilin-3A, SULFATE ION, [(2R)-2-octanoyloxy-3-[oxidanyl-[(1R,2R,3S,4R,5R,6S)-2,3,6-tris(oxidanyl)-4,5-diphosphonooxy-cyclohexyl]oxy-phosphoryl]oxy-propyl] octanoate | | Authors: | Guillen, J, Ferrer-Orta, C, Buxaderas, M, Perez-sanchez, D, Guerrero-Valero, M, Luengo-Gil, G, Pous, J, Guerra, P, Gomez-Fernandez, J.C, Verdaguer, N, Corbalan-Garcia, S. | | Deposit date: | 2013-11-27 | | Release date: | 2013-12-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural insights into the Ca2+ and PI(4,5)P2 binding modes of the C2 domains of rabphilin 3A and synaptotagmin 1.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4NP9
 
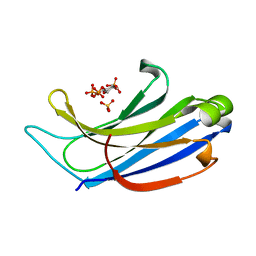 | | Structure of Rabphilin C2A domain bound to IP3 | | Descriptor: | D-MYO-INOSITOL-1,4,5-TRIPHOSPHATE, Rabphilin-3A, SULFATE ION | | Authors: | Guillen, J, Ferrer-Orta, C, Buxaderas, M, Perez-Sanchez, D, Guerrero-Valero, M, Luengo-Gil, G, Pous, J, Guerra, P, Gomez-Fernandez, J.C, Verdaguer, N, Corbalan-Garcia, S. | | Deposit date: | 2013-11-21 | | Release date: | 2013-12-25 | | Last modified: | 2022-08-24 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Structural insights into the Ca2+ and PI(4,5)P2 binding modes of the C2 domains of rabphilin 3A and synaptotagmin 1.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4HW1
 
 | | Multiple Crystal structures of an all-AT DNA dodecamer stabilized by weak interactions. | | Descriptor: | DNA (5'-D(*AP*AP*TP*AP*AP*AP*TP*TP*TP*AP*TP*T)-3'), MAGNESIUM ION | | Authors: | Acosta-Reyes, F, Subirana, J.A, Pous, J, Condom, N, Malinina, L, Campos, J.L. | | Deposit date: | 2012-11-07 | | Release date: | 2013-11-20 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Polymorphic crystal structures of an all-AT DNA dodecamer.
Biopolymers, 103, 2015
|
|
4J2I
 
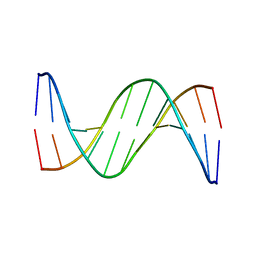 | | Multiple crystal structures of an all-AT DNA dodecamer stabilized by weak interactions | | Descriptor: | 5'-D(*AP*AP*TP*AP*AP*AP*TP*TP*TP*AP*TP*T)-3' | | Authors: | Acosta-Reyes, F.J, Subirana, J.A, Pous, J, Sanchez, R, Condom, N, Baldini, R, Malinina, L, Campos, J.L. | | Deposit date: | 2013-02-04 | | Release date: | 2014-02-05 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.98 Å) | | Cite: | Polymorphic crystal structures of an all-AT DNA dodecamer.
Biopolymers, 103, 2015
|
|
