5M2F
 
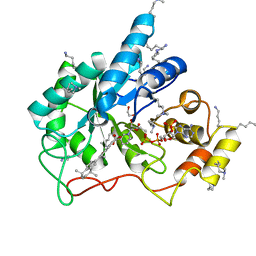 | | Crystal structure of human AKR1B10 complexed with NADP+ and the synthetic retinoid UVI2008 | | Descriptor: | 1,2-ETHANEDIOL, 3-bromo-4-[(1E)-2-(5,5,8,8-tetramethyl-5,6,7,8-tetrahydronaphthalen-2-yl)prop-1-en-1-yl]benzoic acid, Aldo-keto reductase family 1 member B10, ... | | Authors: | Ruiz, F.X, Cousido-Siah, A, Mitschler, A, Porte, S, Alvarez, S, Dominguez, M, Alvarez, R, de Lera, A.R, Pares, X, Farres, J, Podjarny, A. | | Deposit date: | 2016-10-12 | | Release date: | 2017-02-15 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.503 Å) | | Cite: | Structural basis for the inhibition of AKR1B10 by the C3 brominated TTNPB derivative UVI2008.
Chem. Biol. Interact., 276, 2017
|
|
6O9E
 
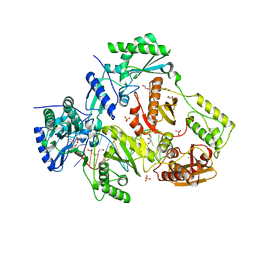 | | Structure of HIV-1 Reverse Transcriptase in complex with DNA and INDOPY-1 | | Descriptor: | 5-methyl-1-(4-nitrophenyl)-2-oxo-2,5-dihydro-1H-pyrido[3,2-b]indole-3-carbonitrile, AMMONIUM ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Ruiz, F.X, Hoang, A, Das, K, Arnold, E. | | Deposit date: | 2019-03-13 | | Release date: | 2019-10-23 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural Basis of HIV-1 Inhibition by Nucleotide-Competing Reverse Transcriptase Inhibitor INDOPY-1.
J.Med.Chem., 62, 2019
|
|
6BHJ
 
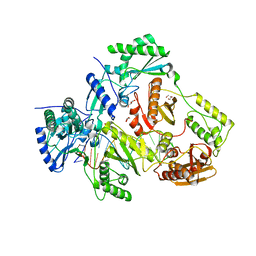 | | Structure of HIV-1 Reverse Transcriptase Bound to a 38-mer Hairpin Template-Primer RNA-DNA Aptamer | | Descriptor: | 38-MER RNA-DNA Aptamer, GLYCEROL, HIV-1 REVERSE TRANSCRIPTASE P51 subunit, ... | | Authors: | Ruiz, F.X, Miller, M.T, Tuske, S, Das, K, Arnold, E. | | Deposit date: | 2017-10-30 | | Release date: | 2018-10-31 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Integrative Structural Biology Studies of HIV-1 Reverse Transcriptase Binding to a High-Affinity DNA Aptamer
Curr Res Struct Biol, 2020
|
|
4XZH
 
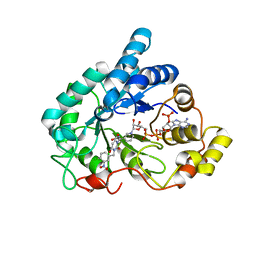 | | Crystal structure of human Aldose Reductase complexed with NADP+ and JF0048 | | Descriptor: | Aldose reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, [3-(4-chloro-3-nitrobenzyl)-2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl]acetic acid | | Authors: | Cousido-Siah, A, Ruiz, F.X, Mitschler, A, Dominguez, M, de Lera, A.R, Farres, J, Pares, X, Podjarny, A. | | Deposit date: | 2015-02-04 | | Release date: | 2015-11-18 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Structural Determinants of the Selectivity of 3-Benzyluracil-1-acetic Acids toward Human Enzymes Aldose Reductase and AKR1B10.
Chemmedchem, 10, 2015
|
|
4XZI
 
 | | Crystal structure of human Aldose Reductase complexed with NADP+ and JF0049 | | Descriptor: | Aldose reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, [2,4-dioxo-3-(2,3,4,5-tetrabromo-6-methoxybenzyl)-3,4-dihydropyrimidin-1(2H)-yl]acetic acid | | Authors: | Cousido-Siah, A, Ruiz, F.X, Mitschler, A, Dominguez, M, de Lera, A.R, Farres, J, Pares, X, Podjarny, A. | | Deposit date: | 2015-02-04 | | Release date: | 2015-11-18 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structural Determinants of the Selectivity of 3-Benzyluracil-1-acetic Acids toward Human Enzymes Aldose Reductase and AKR1B10.
Chemmedchem, 10, 2015
|
|
4XZN
 
 | | Crystal structure of the methylated K125R/V301L AKR1B10 Holoenzyme | | Descriptor: | 1,2-ETHANEDIOL, Aldo-keto reductase family 1 member B10, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Cousido-Siah, A, Ruiz, F.X, Mitschler, A, Podjarny, A. | | Deposit date: | 2015-02-04 | | Release date: | 2015-11-18 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural Determinants of the Selectivity of 3-Benzyluracil-1-acetic Acids toward Human Enzymes Aldose Reductase and AKR1B10.
Chemmedchem, 10, 2015
|
|
4XZL
 
 | | Crystal structure of human AKR1B10 complexed with NADP+ and JF0049 | | Descriptor: | 1,2-ETHANEDIOL, Aldo-keto reductase family 1 member B10, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Cousido-Siah, A, Ruiz, F.X, Mitschler, A, Dominguez, M, de Lera, A.R, Farres, J, Pares, X, Podjarny, A. | | Deposit date: | 2015-02-04 | | Release date: | 2015-11-18 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural Determinants of the Selectivity of 3-Benzyluracil-1-acetic Acids toward Human Enzymes Aldose Reductase and AKR1B10.
Chemmedchem, 10, 2015
|
|
4XZM
 
 | | Crystal structure of the methylated wild-type AKR1B10 holoenzyme | | Descriptor: | 1,2-ETHANEDIOL, Aldo-keto reductase family 1 member B10, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Cousido-Siah, A, Ruiz, F.X, Mitschler, A, Podjarny, A. | | Deposit date: | 2015-02-04 | | Release date: | 2015-11-18 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural Determinants of the Selectivity of 3-Benzyluracil-1-acetic Acids toward Human Enzymes Aldose Reductase and AKR1B10.
Chemmedchem, 10, 2015
|
|
6UL5
 
 | | Crystal structure of HIV-1 reverse transcriptase (RT) in complex with 4-[(4-{4-[(E)-2-cyanoethenyl]-2,6-dimethylphenoxy}thieno[3,2-d]pyrimidin-2-yl)amino]-2-fluorobenzonitrile (24b), a non-nucleoside RT inhibitor | | Descriptor: | 1,2-ETHANEDIOL, 4-[(4-{4-[(E)-2-cyanoethenyl]-2,6-dimethylphenoxy}thieno[3,2-d]pyrimidin-2-yl)amino]-2-fluorobenzonitrile, MAGNESIUM ION, ... | | Authors: | Ruiz, F.X, Pilch, A, Arnold, E. | | Deposit date: | 2019-10-06 | | Release date: | 2020-02-05 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Discovery and Characterization of Fluorine-Substituted Diarylpyrimidine Derivatives as Novel HIV-1 NNRTIs with Highly Improved Resistance Profiles and Low Activity for the hERG Ion Channel.
J.Med.Chem., 63, 2020
|
|
7LRM
 
 | | Structure of HIV-1 Reverse Transcriptase in complex with DNA, dCTP, and CA(2+) ion | | Descriptor: | 2'-DEOXYCYTIDINE-5'-TRIPHOSPHATE, CALCIUM ION, DNA/RNA (38-MER), ... | | Authors: | Hoang, A, Ruiz, F.X, Arnold, E. | | Deposit date: | 2021-02-16 | | Release date: | 2022-02-23 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.14 Å) | | Cite: | Structural basis of HIV inhibition by L-nucleosides: Opportunities for drug development and repurposing.
Drug Discov Today, 27, 2022
|
|
7LRY
 
 | | Structure of HIV-1 Reverse Transcriptase in complex with DNA, (-)FTC-TP, and CA(2+) ion | | Descriptor: | AMMONIUM ION, CALCIUM ION, DNA/RNA (38-MER), ... | | Authors: | Hoang, A, Ruiz, F.X, Arnold, E. | | Deposit date: | 2021-02-17 | | Release date: | 2022-02-23 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structural basis of HIV inhibition by L-nucleosides: Opportunities for drug development and repurposing.
Drug Discov Today, 27, 2022
|
|
7LRI
 
 | | Structure of HIV-1 Reverse Transcriptase in complex with DNA, dTTP, and CA(2+) ion | | Descriptor: | CALCIUM ION, DNA/RNA (38-MER), GLYCEROL, ... | | Authors: | Hoang, A, Ruiz, F.X, Arnold, E. | | Deposit date: | 2021-02-16 | | Release date: | 2022-02-23 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Structural basis of HIV inhibition by L-nucleosides: Opportunities for drug development and repurposing.
Drug Discov Today, 27, 2022
|
|
7LSK
 
 | | Structure of HIV-1 Reverse Transcriptase in complex with DNA, L-dTTP, and CA(2+) ion | | Descriptor: | 1-{2-deoxy-5-O-[(S)-hydroxy{[(S)-hydroxy(phosphonooxy)phosphoryl]oxy}phosphoryl]-beta-L-erythro-pentofuranosyl}-5-methylpyrimidine-2,4(1H,3H)-dione, AMMONIUM ION, CALCIUM ION, ... | | Authors: | Hoang, A, Ruiz, F.X, Arnold, E. | | Deposit date: | 2021-02-18 | | Release date: | 2022-02-23 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis of HIV inhibition by L-nucleosides: Opportunities for drug development and repurposing.
Drug Discov Today, 27, 2022
|
|
7LRX
 
 | | Structure of HIV-1 Reverse Transcriptase in complex with DNA, L-dCTP, and CA(2+) ion | | Descriptor: | 4-amino-1-{2-deoxy-5-O-[(R)-hydroxy{[(S)-hydroxy(phosphonooxy)phosphoryl]oxy}phosphoryl]-beta-L-erythro-pentofuranosyl}pyrimidin-2(1H)-one, AMMONIUM ION, CALCIUM ION, ... | | Authors: | Hoang, A, Ruiz, F.X, Arnold, E. | | Deposit date: | 2021-02-17 | | Release date: | 2022-02-23 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural basis of HIV inhibition by L-nucleosides: Opportunities for drug development and repurposing.
Drug Discov Today, 27, 2022
|
|
7KWU
 
 | | Crystal Structure of HIV-1 RT in Complex with 16c (K07-15) | | Descriptor: | 1,2-ETHANEDIOL, 4-[(4-{[4-(4-cyano-2,6-dimethylphenoxy)-5-(pyridin-4-yl)pyrimidin-2-yl]amino}piperidin-1-yl)methyl]benzamide, MAGNESIUM ION, ... | | Authors: | Ruiz, F.X, Arnold, E. | | Deposit date: | 2020-12-02 | | Release date: | 2021-03-31 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | 2,4,5-Trisubstituted Pyrimidines as Potent HIV-1 NNRTIs: Rational Design, Synthesis, Activity Evaluation, and Crystallographic Studies.
J.Med.Chem., 64, 2021
|
|
4LB4
 
 | | Crystal structure of human AR complexed with NADP+ and {2-[(4-bromo-2,3,5,6-tetrafluorobenzyl)carbamoyl]-5-chlorophenoxy}acetic acid | | Descriptor: | Aldose reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, {2-[(4-bromo-2,3,5,6-tetrafluorobenzyl)carbamoyl]-5-chlorophenoxy}acetic acid | | Authors: | Cousido-Siah, A, Mitschler, A, Ruiz, F.X, Fanfrlik, J, Kolar, M, Hobza, P, Podjarny, A. | | Deposit date: | 2013-06-20 | | Release date: | 2014-04-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (0.8 Å) | | Cite: | Modulation of aldose reductase inhibition by halogen bond tuning.
Acs Chem.Biol., 8, 2013
|
|
4LBS
 
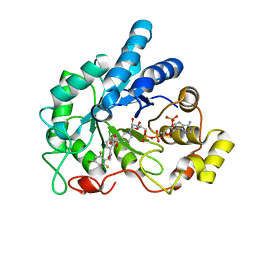 | | Crystal structure of human AR complexed with NADP+ and {2-[(4-bromo-2,6-difluorobenzyl)carbamoyl]-5-chlorophenoxy}acetic acid | | Descriptor: | Aldose reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, {2-[(4-bromo-2,6-difluorobenzyl)carbamoyl]-5-chlorophenoxy}acetic acid | | Authors: | Cousido-Siah, A, Mitschler, A, Ruiz, F.X, Fanfrlik, J, Kolar, M, Hobza, P, Podjarny, A. | | Deposit date: | 2013-06-21 | | Release date: | 2014-04-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (0.76 Å) | | Cite: | Modulation of aldose reductase inhibition by halogen bond tuning.
Acs Chem.Biol., 8, 2013
|
|
4XR8
 
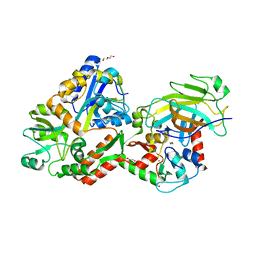 | | Crystal structure of the HPV16 E6/E6AP/p53 ternary complex at 2.25 A resolution | | Descriptor: | 1,2-ETHANEDIOL, Cellular tumor antigen p53, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Martinez-Zapien, D, Ruiz, F.X, Mitschler, A, Podjarny, A, Trave, G, Zanier, K. | | Deposit date: | 2015-01-20 | | Release date: | 2016-02-03 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structure of the E6/E6AP/p53 complex required for HPV-mediated degradation of p53.
Nature, 529, 2016
|
|
1ZUA
 
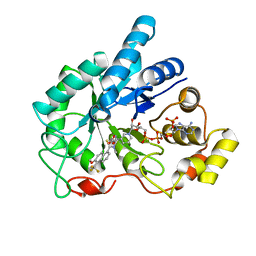 | | Crystal Structure Of AKR1B10 Complexed With NADP+ And Tolrestat | | Descriptor: | Aldo-keto reductase family 1 member B10, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, TOLRESTAT | | Authors: | Gallego, O, Ruiz, F.X, Ardevol, A, Dominguez, M, Alvarez, R, de Lera, A.R, Rovira, C, Farres, J, Fita, I, Pares, X. | | Deposit date: | 2005-05-30 | | Release date: | 2006-11-07 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Structural basis for the high all-trans-retinaldehyde reductase activity of the tumor marker AKR1B10.
Proc.Natl.Acad.Sci.USA, 104, 2007
|
|
4QXI
 
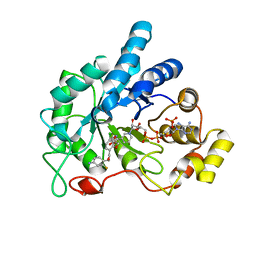 | | Crystal structure of human AR complexed with NADP+ and AK198 | | Descriptor: | Aldose reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, {2-[(4-amino-2-fluorobenzyl)carbamoyl]-5-chlorophenoxy}acetic acid | | Authors: | Cousido-Siah, A, Mitschler, A, Ruiz, F.X, Fanfrlik, J, Hobza, P, Podjarny, A.D. | | Deposit date: | 2014-07-21 | | Release date: | 2015-07-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (0.867 Å) | | Cite: | The Effect of Halogen-to-Hydrogen Bond Substitution on Human Aldose Reductase Inhibition.
Acs Chem.Biol., 10, 2015
|
|
8UOB
 
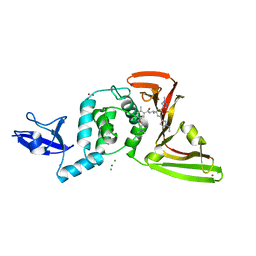 | | SARS-CoV-2 Papain-like protease (PLpro) with Inhibitor Jun12682 | | Descriptor: | 5-[2-(dimethylamino)ethoxy]-N-{(1R)-1-[(3M,5P)-3-(1-ethyl-1H-pyrazol-3-yl)-5-(1-methyl-1H-pyrazol-4-yl)phenyl]ethyl}-2-methylbenzamide, CHLORIDE ION, Papain-like protease nsp3, ... | | Authors: | Ansari, A, Tan, B, Ruiz, F.X, Wang, J, Arnold, E. | | Deposit date: | 2023-10-19 | | Release date: | 2024-04-03 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | Design of a SARS-CoV-2 papain-like protease inhibitor with antiviral efficacy in a mouse model.
Science, 383, 2024
|
|
8UUG
 
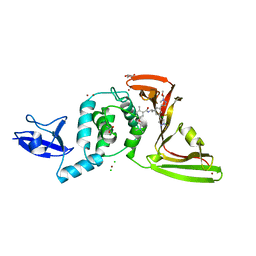 | | SARS-CoV-2 papain-like protease (PLpro) with inhibitor Jun12303 | | Descriptor: | ACETATE ION, CHLORIDE ION, GLYCEROL, ... | | Authors: | Ansari, A, Tan, B, Ruiz, F.X, Arnold, E, Wang, J. | | Deposit date: | 2023-11-01 | | Release date: | 2024-04-03 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.74 Å) | | Cite: | Design of a SARS-CoV-2 papain-like protease inhibitor with antiviral efficacy in a mouse model.
Science, 383, 2024
|
|
8UUH
 
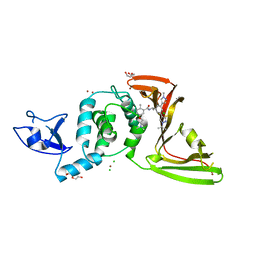 | | SARS-CoV-2 papain-like protease (PLpro) with inhibitor Jun12199 | | Descriptor: | 5-[2-(dimethylamino)ethoxy]-2-methyl-N-[(1R)-1-{(3M,5P)-3-(1-methyl-1H-pyrazol-4-yl)-5-[1-(propan-2-yl)-1H-pyrazol-4-yl]phenyl}ethyl]benzamide, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Ansari, A, Tan, B, Ruiz, F.X, Arnold, E, Wang, J. | | Deposit date: | 2023-11-01 | | Release date: | 2024-04-03 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Design of a SARS-CoV-2 papain-like protease inhibitor with antiviral efficacy in a mouse model.
Science, 383, 2024
|
|
8UUW
 
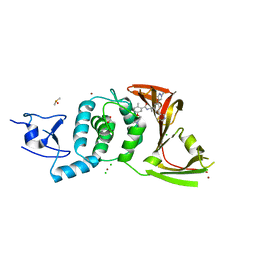 | | SARS-CoV-2 papain-like protease (PLpro) with inhibitor Jun12145 | | Descriptor: | 5-[2-(dimethylamino)ethoxy]-2-methyl-N-{(1R)-1-[(3P,5M)-3-(1-methyl-1H-pyrazol-4-yl)-5-(1,3-thiazol-5-yl)phenyl]ethyl}benzamide, ACETATE ION, CHLORIDE ION, ... | | Authors: | Ansari, A, Tan, B, Ruiz, F.X, Arnold, E, Wang, J. | | Deposit date: | 2023-11-02 | | Release date: | 2024-04-03 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Design of a SARS-CoV-2 papain-like protease inhibitor with antiviral efficacy in a mouse model.
Science, 383, 2024
|
|
8UUY
 
 | | SARS-CoV-2 papain-like protease (PLpro) complex with inhibitor Jun12129 | | Descriptor: | ACETATE ION, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Ansari, A, Tan, B, Ruiz, F.X, Arnold, E, Wang, J. | | Deposit date: | 2023-11-02 | | Release date: | 2024-04-03 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Design of a SARS-CoV-2 papain-like protease inhibitor with antiviral efficacy in a mouse model.
Science, 383, 2024
|
|
