6RXS
 
 | | Crystal structure of CobB Ac3(A76G,Y92A, I131L, V187Y) in complex with H4K16-Acetyl peptide | | Descriptor: | GLYCEROL, Histone H4, NAD-dependent protein deacylase, ... | | Authors: | Spinck, M, Gasper, R, Neumann, H. | | Deposit date: | 2019-06-08 | | Release date: | 2020-04-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.599 Å) | | Cite: | Evolved, Selective Erasers of Distinct Lysine Acylations.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
6RXJ
 
 | | Crystal structure of CobB wt in complex with H4K16-Acetyl peptide | | Descriptor: | Histone H4, NAD-dependent protein deacylase, ZINC ION | | Authors: | Spinck, M, Gasper, R, Neumann, H. | | Deposit date: | 2019-06-08 | | Release date: | 2020-04-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Evolved, Selective Erasers of Distinct Lysine Acylations.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
6RXP
 
 | | Crystal structure of CobB Ac2 (A76G,I131C,V162A) in complex with H4K16-Crotonyl peptide | | Descriptor: | Histone H4, NAD-dependent protein deacylase, ZINC ION | | Authors: | Spinck, M, Gasper, R, Neumann, H. | | Deposit date: | 2019-06-08 | | Release date: | 2020-04-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Evolved, Selective Erasers of Distinct Lysine Acylations.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
6RXL
 
 | | Crystal structure of CobB wt in complex with H4K16-Crotonyl peptide | | Descriptor: | Histone H4, NAD-dependent protein deacylase, ZINC ION | | Authors: | Spinck, M, Gasper, R, Neumann, H. | | Deposit date: | 2019-06-08 | | Release date: | 2020-04-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Evolved, Selective Erasers of Distinct Lysine Acylations.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
6RXR
 
 | | Crystal structure of CobB Ac2 (A76G, I131C, V162G) in complex with H4K16Cr-2'OH-ADPr peptide intermediate after co-crystallisation | | Descriptor: | Histone H4, NAD-dependent protein deacylase, [[(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl] [(2~{R},3~{R},4~{R},5~{S})-4-[(~{E})-but-2-enoxy]-3,5-bis(oxidanyl)oxolan-2-yl]methyl hydrogen phosphate | | Authors: | Spinck, M, Gasper, R, Neumann, H. | | Deposit date: | 2019-06-08 | | Release date: | 2020-04-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Evolved, Selective Erasers of Distinct Lysine Acylations.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
6RXO
 
 | | Crystal structure of CobB Ac2 (A76G, I131C, V162A) in complex with H4K16-Buturyl peptide | | Descriptor: | Histone H4, NAD-dependent protein deacylase, ZINC ION | | Authors: | Spinck, M, Gasper, R, Neumann, H. | | Deposit date: | 2019-06-08 | | Release date: | 2020-04-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Evolved, Selective Erasers of Distinct Lysine Acylations.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
4BN5
 
 | | Structure of human SIRT3 in complex with SRT1720 inhibitor | | Descriptor: | CARBA-NICOTINAMIDE-ADENINE-DINUCLEOTIDE, GLYCEROL, N-{2-[3-(piperazin-1-ylmethyl)imidazo[2,1-b][1,3]thiazol-6-yl]phenyl}quinoxaline-2-carboxamide, ... | | Authors: | Nguyen, G.T.T, Schaefer, S, Gertz, M, Weyand, M, Steegborn, C. | | Deposit date: | 2013-05-13 | | Release date: | 2013-06-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Structures of Human Sirtuin 3 Complexes with Adp-Ribose and with Carba-Nad+ and Srt1720: Binding Details and Inhibition Mechanism
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4BVG
 
 | | CRYSTAL STRUCTURE OF HUMAN SIRT3 IN COMPLEX WITH NATIVE ALKYLIMIDATE FORMED FROM ACETYL-LYSINE ACS2-PEPTIDE CRYSTALLIZED IN PRESENCE OF THE INHIBITOR EX-527 | | Descriptor: | (2R,3R,4S,5R)-5-({[(R)-{[(R)-{[(2R,3S,4R,5R)-5-(6-AMINO-9H-PURIN-9-YL)-3,4-DIHYDROXYTETRAHYDROFURAN-2-YL]METHOXY}(HYDROXY)PHOSPHORYL]OXY}(HYDROXY)PHOSPHORYL]OXY}METHYL)-3,4-DIHYDROXYTETRAHYDROFURAN-2-YL ACETATE, 1,2-ETHANEDIOL, ACETYL-COENZYME A SYNTHETASE 2-LIKE, ... | | Authors: | Nguyen, G.T.T, Gertz, M, Weyand, M, Steegborn, C. | | Deposit date: | 2013-06-25 | | Release date: | 2013-07-17 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Ex-527 inhibits Sirtuins by exploiting their unique NAD+-dependent deacetylation mechanism.
Proc. Natl. Acad. Sci. U.S.A., 110, 2013
|
|
4BUZ
 
 | | SIR2 COMPLEX STRUCTURE MIXTURE OF EX-527 INHIBITOR AND REACTION PRODUCTS OR OF REACTION SUBSTRATES P53 PEPTIDE AND NAD | | Descriptor: | (1S)-6-chloro-2,3,4,9-tetrahydro-1H-carbazole-1- carboxamide, 1,2-ETHANEDIOL, 2'-O-ACETYL ADENOSINE-5-DIPHOSPHORIBOSE, ... | | Authors: | Weyand, M, Lakshminarasimhan, M, Gertz, M, Steegborn, C. | | Deposit date: | 2013-06-24 | | Release date: | 2013-07-17 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Ex-527 Inhibits Sirtuins by Exploiting Their Unique Nad+-Dependent Deacetylation Mechanism
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4C7B
 
 | | Complex of human Sirt3 with Bromo-Resveratrol and Fluor-De-Lys peptide | | Descriptor: | 5-[(E)-2-(4-bromophenyl)ethenyl]benzene-1,3-diol, GLYCEROL, ISOPROPYL ALCOHOL, ... | | Authors: | Nguyen, G.T.T, Gertz, M, Weyand, M, Steegborn, C. | | Deposit date: | 2013-09-20 | | Release date: | 2013-11-20 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structures of Sirt3 Complexes with 4'-Bromo-Resveratrol Reveal Binding Sites and Inhibition Mechanism.
Chem.Biol., 20, 2013
|
|
4BV2
 
 | | CRYSTAL STRUCTURE OF SIR2 IN COMPLEX WITH THE INHIBITOR EX-527, 2'-O-ACETYL-ADP-RIBOSE AND DEACETYLATED P53-PEPTIDE | | Descriptor: | (1S)-6-chloro-2,3,4,9-tetrahydro-1H-carbazole-1- carboxamide, 2'-O-ACETYL ADENOSINE-5-DIPHOSPHORIBOSE, CELLULAR TUMOR ANTIGEN P53, ... | | Authors: | Gertz, M, Weyand, M, Steegborn, C. | | Deposit date: | 2013-06-24 | | Release date: | 2013-07-17 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Ex-527 Inhibits Sirtuins by Exploiting Their Unique Nad+-Dependent Deacetylation Mechanism
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4BN4
 
 | | Structure of human SIRT3 in complex with ADP-ribose | | Descriptor: | 2-[2-[2-[2-[2-(2-hydroxyethyloxy)ethoxy]ethoxy]ethoxy]ethoxy]ethanoic acid, NAD-DEPENDENT PROTEIN DEACETYLASE SIRTUIN-3, MITOCHONDRIAL, ... | | Authors: | Nguyen, G.T.T, Schaefer, S, Gertz, M, Weyand, M, Steegborn, C. | | Deposit date: | 2013-05-13 | | Release date: | 2013-06-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structures of Human Sirtuin 3 Complexes with Adp-Ribose and with Carba-Nad+ and Srt1720: Binding Details and Inhibition Mechanism
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4BVE
 
 | | CRYSTAL STRUCTURE OF HUMAN SIRT3 IN COMPLEX WITH THIOALKYLIMIDATE FORMED FROM THIO-ACETYL-LYSINE ACS2-PEPTIDE | | Descriptor: | 1,2-ETHANEDIOL, ACETYL-COENZYME A SYNTHETASE 2-LIKE, MITOCHONDRIAL, ... | | Authors: | Gertz, M, Weyand, M, Steegborn, C. | | Deposit date: | 2013-06-25 | | Release date: | 2013-07-17 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Ex-527 Inhibits Sirtuins by Exploiting Their Unique Nad+-Dependent Deacetylation Mechanism
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4BVB
 
 | | CRYSTAL STRUCTURE OF HUMAN SIRT3 IN COMPLEX WITH THE INHIBITOR EX-527 AND ADP-RIBOSE | | Descriptor: | (1S)-6-chloro-2,3,4,9-tetrahydro-1H-carbazole-1- carboxamide, NAD-DEPENDENT PROTEIN DEACETYLASE SIRTUIN-3, MITOCHONDRIAL, ... | | Authors: | Gertz, M, Weyand, M, Steegborn, C. | | Deposit date: | 2013-06-25 | | Release date: | 2013-07-17 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Ex-527 Inhibits Sirtuins by Exploiting Their Unique Nad+-Dependent Deacetylation Mechanism
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4BV3
 
 | | CRYSTAL STRUCTURE OF SIRT3 IN COMPLEX WITH THE INHIBITOR EX-527 AND NAD | | Descriptor: | (1S)-6-chloro-2,3,4,9-tetrahydro-1H-carbazole-1- carboxamide, ADENOSINE-5-DIPHOSPHORIBOSE, CHLORIDE ION, ... | | Authors: | Gertz, M, Nguyen, N.T.T, Weyand, M, Steegborn, C. | | Deposit date: | 2013-06-24 | | Release date: | 2013-07-17 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Ex-527 Inhibits Sirtuins by Exploiting Their Unique Nad+-Dependent Deacetylation Mechanism
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4BVF
 
 | | CRYSTAL STRUCTURE OF HUMAN SIRT3 IN COMPLEX WITH THIOALKYLIMIDATE FORMED FROM THIO-ACETYL-LYSINE ACS2-PEPTIDE CRYSTALLIZED IN PRESENCE OF THE INHIBITOR EX-527 | | Descriptor: | 1,2-ETHANEDIOL, ACETYL-COENZYME A SYNTHETASE 2-LIKE, MITOCHONDRIAL, ... | | Authors: | Gertz, M, Weyand, M, Steegborn, C. | | Deposit date: | 2013-06-25 | | Release date: | 2013-07-17 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Ex-527 Inhibits Sirtuins by Exploiting Their Unique Nad+-Dependent Deacetylation Mechanism
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4C78
 
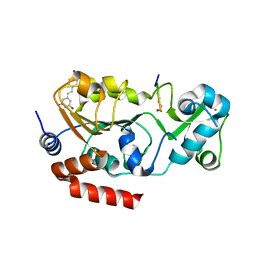 | | Complex of human Sirt3 with Bromo-Resveratrol and ACS2 peptide | | Descriptor: | 5-[(E)-2-(4-bromophenyl)ethenyl]benzene-1,3-diol, ACETYL-COENZYME A SYNTHETASE 2-LIKE, MITOCHONDRIAL, ... | | Authors: | Nguyen, G.T.T, Gertz, M, Weyand, M, Steegborn, C. | | Deposit date: | 2013-09-19 | | Release date: | 2013-11-20 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structures of Sirt3 Complexes with 4'-Bromo-Resveratrol Reveal Binding Sites and Inhibition Mechanism.
Chem.Biol., 20, 2013
|
|
4BVH
 
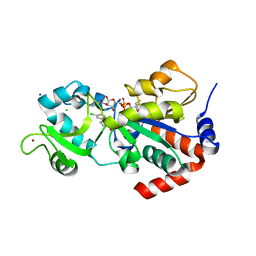 | | CRYSTAL STRUCTURE OF HUMAN SIRT3 IN COMPLEX WITH THE INHIBITOR EX-527 AND 2'-O-ACETYL-ADP-RIBOSE | | Descriptor: | (1S)-6-chloro-2,3,4,9-tetrahydro-1H-carbazole-1- carboxamide, 1,2-ETHANEDIOL, 2'-O-ACETYL ADENOSINE-5-DIPHOSPHORIBOSE, ... | | Authors: | Gertz, M, Weyand, M, Steegborn, C. | | Deposit date: | 2013-06-25 | | Release date: | 2013-07-17 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Ex-527 Inhibits Sirtuins by Exploiting Their Unique Nad+-Dependent Deacetylation Mechanism
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4F4U
 
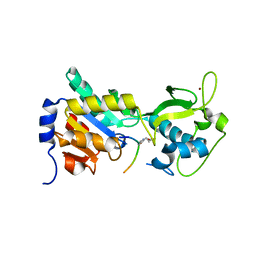 | |
4F56
 
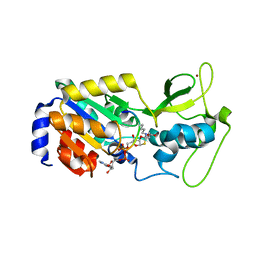 | | The bicyclic intermediate structure provides insights into the desuccinylation mechanism of SIRT5 | | Descriptor: | 3-[(2R,3aR,5R,6R,6aR)-5-({[(S)-{[(S)-{[(2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-3,4-dihydroxytetrahydrofuran-2-yl]methoxy}(hydroxy)phosphoryl]oxy}(hydroxy)phosphoryl]oxy}methyl)-2,6-dihydroxytetrahydrofuro[2,3-d][1,3]oxathiol-2-yl]propanoic acid, NAD-dependent lysine demalonylase and desuccinylase sirtuin-5, mitochondrial, ... | | Authors: | Zhou, Y, Hao, Q. | | Deposit date: | 2012-05-11 | | Release date: | 2012-06-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The Bicyclic Intermediate Structure Provides Insights into the Desuccinylation Mechanism of Human Sirtuin 5 (SIRT5)
J.Biol.Chem., 287, 2012
|
|
4FZ3
 
 | | Crystal structure of SIRT3 in complex with acetyl p53 peptide coupled with 4-amino-7-methylcoumarin | | Descriptor: | NAD-dependent protein deacetylase sirtuin-3, mitochondrial, ZINC ION, ... | | Authors: | Liu, D, Wu, J, Zhang, D, Chen, K, Jiang, H, Liu, H. | | Deposit date: | 2012-07-06 | | Release date: | 2013-03-20 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Discovery and Mechanism Study of SIRT1 Activators that Promote the Deacetylation of Fluorophore-Labeled Substrate
J.Med.Chem., 56, 2013
|
|
4FVT
 
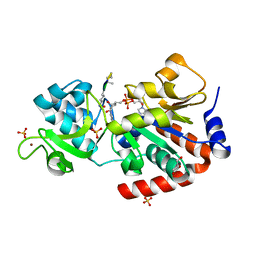 | | Human SIRT3 bound to Ac-ACS peptide and Carba-NAD | | Descriptor: | Acetylated ACS2 peptide, CARBA-NICOTINAMIDE-ADENINE-DINUCLEOTIDE, GLYCEROL, ... | | Authors: | Dai, H. | | Deposit date: | 2012-06-29 | | Release date: | 2012-08-15 | | Last modified: | 2012-09-19 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | Synthesis of Carba-NAD and the Structures of Its Ternary Complexes with SIRT3 and SIRT5.
J.Org.Chem., 77, 2012
|
|
4G1C
 
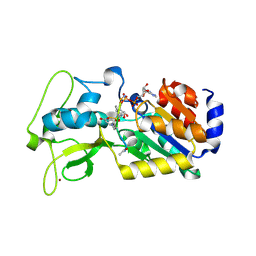 | | Human SIRT5 bound to Succ-IDH2 and Carba-NAD | | Descriptor: | CARBA-NICOTINAMIDE-ADENINE-DINUCLEOTIDE, NAD-dependent protein deacylase sirtuin-5, mitochondrial, ... | | Authors: | Dai, H. | | Deposit date: | 2012-07-10 | | Release date: | 2012-08-15 | | Last modified: | 2012-09-19 | | Method: | X-RAY DIFFRACTION (1.944 Å) | | Cite: | Synthesis of Carba-NAD and the Structures of Its Ternary Complexes with SIRT3 and SIRT5.
J.Org.Chem., 77, 2012
|
|
4HDA
 
 | |
4HD8
 
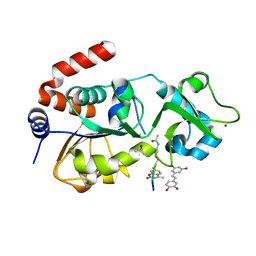 | | Crystal structure of human Sirt3 in complex with Fluor-de-Lys peptide and piceatannol | | Descriptor: | Fluor-de-Lys peptide, ISOPROPYL ALCOHOL, NAD-dependent protein deacetylase sirtuin-3, ... | | Authors: | Nguyen, G.T.T, Gertz, M, Steegborn, C. | | Deposit date: | 2012-10-02 | | Release date: | 2012-12-05 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A molecular mechanism for direct sirtuin activation by resveratrol.
Plos One, 7, 2012
|
|
