5VSK
 
 | | Structure of DUB complex | | Descriptor: | 7-chloro-3-({4-hydroxy-1-[(3S)-3-phenylbutanoyl]piperidin-4-yl}methyl)quinazolin-4(3H)-one, Ubiquitin carboxyl-terminal hydrolase 7, ZINC ION | | Authors: | Seo, H.-Y, Dhe-Paganon, S. | | Deposit date: | 2017-05-11 | | Release date: | 2017-12-20 | | Last modified: | 2018-01-03 | | Method: | X-RAY DIFFRACTION (3.33 Å) | | Cite: | Structure-Guided Development of a Potent and Selective Non-covalent Active-Site Inhibitor of USP7.
Cell Chem Biol, 24, 2017
|
|
1W6V
 
 | | Solution structure of the DUSP domain of hUSP15 | | Descriptor: | UBIQUITIN CARBOXYL-TERMINAL HYDROLASE 15 | | Authors: | De Jong, R.D, Ab, E, Diercks, T, Truffault, V, Daniels, M, Kaptein, R, Folkers, G.E. | | Deposit date: | 2004-08-24 | | Release date: | 2006-01-12 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the Human Ubiquitin-Specific Protease 15 Dusp Domain.
J.Biol.Chem., 281, 2006
|
|
5NGE
 
 | | Crystal structure of USP7 in complex with the non-covalent inhibitor, FT671 | | Descriptor: | 5-[[1-[(3~{S})-4,4-bis(fluoranyl)-3-(3-fluoranylpyrazol-1-yl)butanoyl]-4-oxidanyl-piperidin-4-yl]methyl]-1-(4-fluorophenyl)pyrazolo[3,4-d]pyrimidin-4-one, Ubiquitin carboxyl-terminal hydrolase 7 | | Authors: | Turnbull, A.P, Krajewski, W.W, Ioannidis, S, Kessler, B.M, Komander, D. | | Deposit date: | 2017-03-17 | | Release date: | 2017-10-18 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Molecular basis of USP7 inhibition by selective small-molecule inhibitors.
Nature, 550, 2017
|
|
5NGF
 
 | | Crystal structure of USP7 in complex with the covalent inhibitor, FT827 | | Descriptor: | 1,2-ETHANEDIOL, Ubiquitin carboxyl-terminal hydrolase 7, ~{N}-[2-[4-[4-[(1-methyl-4-oxidanylidene-pyrazolo[3,4-d]pyrimidin-5-yl)methyl]-4-oxidanyl-piperidin-1-yl]carbonylphenyl]phenyl]ethanesulfonamide | | Authors: | Krajewski, W.W, Turnbull, A.P, Ioannidis, S, Kessler, B.M, Komander, D. | | Deposit date: | 2017-03-17 | | Release date: | 2017-10-18 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Molecular basis of USP7 inhibition by selective small-molecule inhibitors.
Nature, 550, 2017
|
|
5CHV
 
 | | Crystal structure of USP18-ISG15 complex | | Descriptor: | CHLORIDE ION, SULFATE ION, Ubiquitin-like protein ISG15, ... | | Authors: | Fritz, G, Basters, A. | | Deposit date: | 2015-07-10 | | Release date: | 2016-09-28 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.005 Å) | | Cite: | Structural basis of the specificity of USP18 toward ISG15.
Nat. Struct. Mol. Biol., 24, 2017
|
|
5CHT
 
 | | Crystal structure of USP18 | | Descriptor: | Ubl carboxyl-terminal hydrolase 18, ZINC ION | | Authors: | Fritz, G, Basters, A. | | Deposit date: | 2015-07-10 | | Release date: | 2016-06-29 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis of the specificity of USP18 toward ISG15.
Nat. Struct. Mol. Biol., 24, 2017
|
|
5K1A
 
 | |
5K1C
 
 | | Crystal structure of the UAF1/WDR20/USP12 complex | | Descriptor: | PHOSPHATE ION, TRIS(HYDROXYETHYL)AMINOMETHANE, Ubiquitin carboxyl-terminal hydrolase 12, ... | | Authors: | Li, H, D'Andrea, A.D, Zheng, N. | | Deposit date: | 2016-05-18 | | Release date: | 2016-07-20 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Allosteric Activation of Ubiquitin-Specific Proteases by beta-Propeller Proteins UAF1 and WDR20.
Mol.Cell, 63, 2016
|
|
5K16
 
 | |
5K1B
 
 | |
6GHA
 
 | | USP15 catalytic domain structure | | Descriptor: | Ubiquitin carboxyl-terminal hydrolase 15,Ubiquitin carboxyl-terminal hydrolase 15, ZINC ION | | Authors: | Ward, S.J, Gratton, H.E, Caulton, S.G, Emsley, J, Dreveny, I. | | Deposit date: | 2018-05-06 | | Release date: | 2018-09-26 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | The structure of the deubiquitinase USP15 reveals a misaligned catalytic triad and an open ubiquitin-binding channel.
J. Biol. Chem., 293, 2018
|
|
6GH9
 
 | | USP15 catalytic domain in complex with small molecule | | Descriptor: | 1,4-DIHYDROXY-5,8-BIS({2-[(2-HYDROXYETHYL)AMINO]ETHYL}AMINO)-9,10-ANTHRACENEDIONE, DIMETHYL SULFOXIDE, Ubiquitin carboxyl-terminal hydrolase 15, ... | | Authors: | Ward, S.J, Gratton, H.E, Caulton, S.G, Emsley, J, Dreveny, I. | | Deposit date: | 2018-05-06 | | Release date: | 2018-09-26 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | The structure of the deubiquitinase USP15 reveals a misaligned catalytic triad and an open ubiquitin-binding channel.
J. Biol. Chem., 293, 2018
|
|
5GVI
 
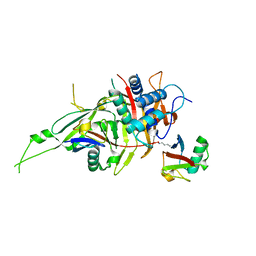 | | Zebrafish USP30 in complex with Lys6-linked diubiquitin | | Descriptor: | Ubiquitin carboxyl-terminal hydrolase 30, ZINC ION, ubiquitin | | Authors: | Sato, Y, Fukai, S. | | Deposit date: | 2016-09-05 | | Release date: | 2017-09-13 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Structural basis for specific cleavage of Lys6-linked polyubiquitin chains by USP30
Nat. Struct. Mol. Biol., 24, 2017
|
|
5CVL
 
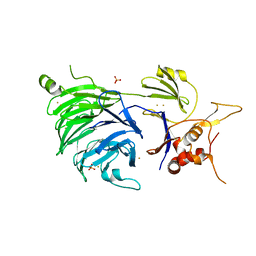 | | WDR48 (UAF-1), residues 2-580 | | Descriptor: | GOLD ION, PHOSPHATE ION, WD repeat-containing protein 48 | | Authors: | HARRIS, S.F, YIN, J. | | Deposit date: | 2015-07-27 | | Release date: | 2015-10-07 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural Insights into WD-Repeat 48 Activation of Ubiquitin-Specific Protease 46.
Structure, 23, 2015
|
|
8OYP
 
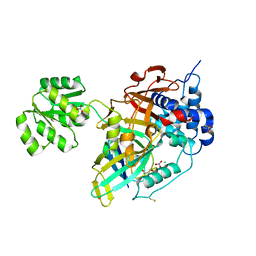 | | Crystal structure of Ubiquitin specific protease 11 (USP11) in complex with a substrate mimetic | | Descriptor: | CADMIUM ION, CHLORIDE ION, GLYCEROL, ... | | Authors: | Maurer, S.K, Caulton, S.G, Ward, S.J, Emsley, J, Dreveny, I. | | Deposit date: | 2023-05-05 | | Release date: | 2023-10-18 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Ubiquitin-specific protease 11 structure in complex with an engineered substrate mimetic reveals a molecular feature for deubiquitination selectivity.
J.Biol.Chem., 299, 2023
|
|
6KAC
 
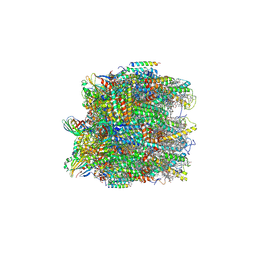 | | Cryo-EM structure of the C2S2-type PSII-LHCII supercomplex from Chlamydomonas reihardtii | | Descriptor: | (1R,3R)-6-{(3E,5E,7E,9E,11E,13E,15E,17E)-18-[(1S,4R,6R)-4-HYDROXY-2,2,6-TRIMETHYL-7-OXABICYCLO[4.1.0]HEPT-1-YL]-3,7,12,16-TETRAMETHYLOCTADECA-1,3,5,7,9,11,13,15,17-NONAENYLIDENE}-1,5,5-TRIMETHYLCYCLOHEXANE-1,3-DIOL, (3R,3'R,6S)-4,5-DIDEHYDRO-5,6-DIHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, (3S,5R,6S,3'S,5'R,6'S)-5,6,5',6'-DIEPOXY-5,6,5',6'- TETRAHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, ... | | Authors: | Sheng, X, Li, A.J, Song, D.F, Liu, Z.F. | | Deposit date: | 2019-06-21 | | Release date: | 2019-10-23 | | Last modified: | 2019-12-25 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structural insight into light harvesting for photosystem II in green algae.
Nat.Plants, 5, 2019
|
|
4MSX
 
 | | Crystal structure of an essential yeast splicing factor | | Descriptor: | ACETATE ION, Pre-mRNA-splicing factor SAD1, ZINC ION | | Authors: | Hadjivassiliou, H, Guthrie, C, Rosenberg, O.S. | | Deposit date: | 2013-09-18 | | Release date: | 2014-04-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | The crystal structure of S. cerevisiae Sad1, a catalytically inactive deubiquitinase that is broadly required for pre-mRNA splicing.
Rna, 20, 2014
|
|
5QYE
 
 | | PanDDA analysis group deposition -- Aar2/RNaseH in complex with fragment F2X-Entry E12a | | Descriptor: | (2R)-6-amino-3-methyl-2,3-dihydro-1,3-benzoxazol-2-ol, A1 cistron-splicing factor AAR2, Pre-mRNA-splicing factor 8, ... | | Authors: | Weiss, M.S, Wollenhaupt, J, Metz, A, Barthel, T, Lima, G.M.A, Heine, A, Mueller, U, Klebe, G. | | Deposit date: | 2020-02-12 | | Release date: | 2020-06-10 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | F2X-Universal and F2X-Entry: Structurally Diverse Compound Libraries for Crystallographic Fragment Screening.
Structure, 28, 2020
|
|
8SY2
 
 | |
8SY3
 
 | |
1R0N
 
 | | Crystal Structure of Heterodimeric Ecdsyone receptor DNA binding complex | | Descriptor: | Ecdsyone Response Element, Ecdysone Response Element, Ecdysone receptor, ... | | Authors: | Devarakonda, S, Harp, J.M, Kim, Y, Ozyhar, A, Rastinejad, F. | | Deposit date: | 2003-09-22 | | Release date: | 2003-10-21 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of the heterodimeric Ecdysone Receptor DNA-binding complex
Embo J., 22, 2003
|
|
4WA6
 
 | |
8XPN
 
 | | The Crystal Structure of USP8 from Biortus. | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, Ubiquitin carboxyl-terminal hydrolase 8, ... | | Authors: | Wang, F, Cheng, W, Yuan, Z, Lin, D, Wang, J. | | Deposit date: | 2024-01-04 | | Release date: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The Crystal Structure of USP8 from Biortus.
To Be Published
|
|
4W4U
 
 | |
1NB8
 
 | | Structure of the catalytic domain of USP7 (HAUSP) | | Descriptor: | Ubiquitin carboxyl-terminal hydrolase 7 | | Authors: | Hu, M, Li, P, Li, M, Li, W, Yao, T, Wu, J.-W, Gu, W, Cohen, R.E, Shi, Y. | | Deposit date: | 2002-12-02 | | Release date: | 2003-01-07 | | Last modified: | 2018-04-04 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of a UBP-family deubiquitinating enzyme in isolation and in complex with ubiquitin aldehyde
Cell(Cambridge,Mass.), 111, 2002
|
|
