6VR1
 
 | | Complex of HLA-A2, a class I MHC, with a p53 peptide | | Descriptor: | Beta-2-microglobulin, Cellular tumor antigen p53 peptide, MHC class I antigen | | Authors: | Wu, D, Pierce, B.G, Gallagher, D.T, Mariuzza, R.A. | | Deposit date: | 2020-02-06 | | Release date: | 2020-06-10 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Structural basis for oligoclonal T cell recognition of a shared p53 cancer neoantigen.
Nat Commun, 11, 2020
|
|
6VRN
 
 | | T cell receptor-p53-HLA-A2 complex | | Descriptor: | Beta-2-microglobulin, Cellular tumor antigen p53 peptide, MHC class I antigen, ... | | Authors: | Wu, D, Gallagher, D.T, Gowthaman, R, Pierce, B.G, Mariuzza, R.A. | | Deposit date: | 2020-02-08 | | Release date: | 2020-06-17 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | Structural basis for oligoclonal T cell recognition of a shared p53 cancer neoantigen.
Nat Commun, 11, 2020
|
|
6VRM
 
 | | T cell receptor-p53-HLA-A2 complex | | Descriptor: | Beta-2-microglobulin, Cellular tumor antigen p53 peptide, MHC class I antigen, ... | | Authors: | Wu, D, Gallagher, D.T, Gowthaman, R, Pierce, B.G, Mariuzza, R.A. | | Deposit date: | 2020-02-08 | | Release date: | 2020-06-17 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Structural basis for oligoclonal T cell recognition of a shared p53 cancer neoantigen.
Nat Commun, 11, 2020
|
|
6VR5
 
 | | Complex of HLA-A2, a class I MHC, with a p53 peptide | | Descriptor: | Beta-2-microglobulin, Cellular tumor antigen p53 peptide, MHC class I antigen | | Authors: | Wu, D, Gallagher, D.T, Pierce, B.G, Mariuzza, R.A. | | Deposit date: | 2020-02-06 | | Release date: | 2020-06-17 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Structural basis for oligoclonal T cell recognition of a shared p53 cancer neoantigen.
Nat Commun, 11, 2020
|
|
4HJE
 
 | | Crystal structure of p53 core domain in complex with DNA | | Descriptor: | Cellular tumor antigen p53, DNA (5'-D(*AP*GP*GP*CP*TP*TP*GP*TP*CP*TP*CP*TP*AP*AP*CP*TP*TP*GP*TP*GP*A)-3'), DNA (5'-D(*TP*CP*AP*CP*AP*AP*GP*TP*TP*AP*GP*AP*GP*AP*CP*AP*AP*GP*CP*CP*T)-3'), ... | | Authors: | Chen, Y, Chen, L. | | Deposit date: | 2012-10-12 | | Release date: | 2013-07-17 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.907 Å) | | Cite: | Structure of p53 binding to the BAX response element reveals DNA unwinding and compression to accommodate base-pair insertion.
Nucleic Acids Res., 41, 2013
|
|
7DHY
 
 | | Arsenic-bound p53 DNA-binding domain mutant G245S | | Descriptor: | ARSENIC, Cellular tumor antigen p53, GLYCEROL, ... | | Authors: | Chen, S, Lu, M. | | Deposit date: | 2020-11-18 | | Release date: | 2021-03-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Arsenic Trioxide Rescues Structural p53 Mutations through a Cryptic Allosteric Site.
Cancer Cell, 39, 2021
|
|
7DHZ
 
 | | Arsenic-bound p53 DNA-binding domain mutant R249S | | Descriptor: | ARSENIC, Cellular tumor antigen p53, GLYCEROL, ... | | Authors: | Chen, S, Lu, M. | | Deposit date: | 2020-11-18 | | Release date: | 2021-03-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Arsenic Trioxide Rescues Structural p53 Mutations through a Cryptic Allosteric Site.
Cancer Cell, 39, 2021
|
|
6IJQ
 
 | | Solution structure of BCL-XL bound to P73-TAD peptide | | Descriptor: | Bcl-2-like protein 1,Bcl-2-like protein 1, Tumor protein p73 | | Authors: | Yoon, M.-K, Ha, J.-H, Lee, M.-S, Chi, S.-W. | | Deposit date: | 2018-10-11 | | Release date: | 2018-11-21 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Cytoplasmic pro-apoptotic function of the tumor suppressor p73 is mediated through a modified mode of recognition of the anti-apoptotic regulator Bcl-XL.
J. Biol. Chem., 293, 2018
|
|
6GGF
 
 | |
3ZME
 
 | |
2MEJ
 
 | |
5AOI
 
 | |
5AOM
 
 | | Structure of the p53 cancer mutant Y220C with bound small molecule PhiKan883 | | Descriptor: | CELLULAR TUMOR ANTIGEN P53, GLYCEROL, N-(5-chloranyl-2-oxidanyl-phenyl)piperidine-4-carboxamide, ... | | Authors: | Joerger, A.C, Boeckler, F.M, Wilcken, R. | | Deposit date: | 2015-09-10 | | Release date: | 2015-12-16 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Exploiting Transient Protein States for the Design of Small-Molecule Stabilizers of Mutant P53.
Structure, 23, 2015
|
|
5AOL
 
 | |
5AOJ
 
 | | Structure of the p53 cancer mutant Y220C in complex with 2-hydroxy-3, 5-diiodo-4-(1H-pyrrol-1-yl)benzoic acid | | Descriptor: | 2-hydroxy-3,5-diiodo-4-(1H-pyrrol-1-yl)benzoic acid, CELLULAR TUMOR ANTIGEN P53, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Joerger, A.C, Baud, M.G, Bauer, M.R, Fersht, A.R. | | Deposit date: | 2015-09-10 | | Release date: | 2015-12-16 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Exploiting Transient Protein States for the Design of Small-Molecule Stabilizers of Mutant P53.
Structure, 23, 2015
|
|
5AOK
 
 | | Structure of the p53 cancer mutant Y220C with bound small molecule PhiKan7099 | | Descriptor: | 5-[2-cyclopropyl-5-(1H-pyrrol-1-yl)-1,3-oxazol-4-yl]-1H-1,2,3,4-tetrazole, CELLULAR TUMOR ANTIGEN P53, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Joerger, A.C. | | Deposit date: | 2015-09-10 | | Release date: | 2015-12-16 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Exploiting Transient Protein States for the Design of Small-Molecule Stabilizers of Mutant P53.
Structure, 23, 2015
|
|
5AB9
 
 | |
5ABA
 
 | |
2LY4
 
 | | HMGB1-facilitated p53 DNA binding occurs via HMG-box/p53 transactivation domain interaction and is regulated by the acidic tail | | Descriptor: | Cellular tumor antigen p53, High mobility group protein B1 | | Authors: | Rowell, J.P, Simpson, K.L, Stott, K, Watson, M, Thomas, J.O. | | Deposit date: | 2012-09-12 | | Release date: | 2012-10-31 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | HMGB1-Facilitated p53 DNA Binding Occurs via HMG-Box/p53 Transactivation Domain Interaction, Regulated by the Acidic Tail.
Structure, 20, 2012
|
|
6T58
 
 | | Structure determination of the transactivation domain of p53 in complex with S100A4 using annexin A2 as a crystallization chaperone | | Descriptor: | CALCIUM ION, Cellular tumor antigen p53,Protein S100-A4,Protein S100-A4,Annexin A2, GLYCEROL | | Authors: | Ecsedi, P, Gogl, G, Nyitray, L. | | Deposit date: | 2019-10-15 | | Release date: | 2020-05-27 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structure Determination of the Transactivation Domain of p53 in Complex with S100A4 Using Annexin A2 as a Crystallization Chaperone.
Structure, 28, 2020
|
|
8F2I
 
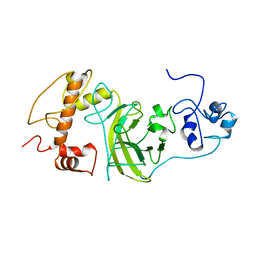 | | P53 monomer structure | | Descriptor: | Cellular tumor antigen p53 | | Authors: | Solares, M, Kelly, D.F. | | Deposit date: | 2022-11-08 | | Release date: | 2022-11-23 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (5 Å) | | Cite: | High-Resolution Imaging of Human Cancer Proteins Using Microprocessor Materials.
Chembiochem, 23, 2022
|
|
2PCX
 
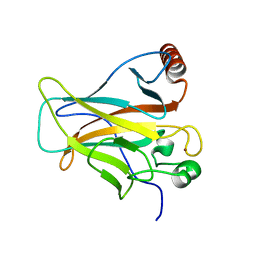 | | Crystal structure of p53DBD(R282Q) at 1.54-angstrom Resolution | | Descriptor: | Cellular tumor antigen p53, ZINC ION | | Authors: | Tu, C, Shaw, G, Ji, X. | | Deposit date: | 2007-03-30 | | Release date: | 2008-04-08 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Impact of low-frequency hotspot mutation R282Q on the structure of p53 DNA-binding domain as revealed by crystallography at 1.54 angstroms resolution.
Acta Crystallogr.,Sect.D, 64, 2008
|
|
2MWO
 
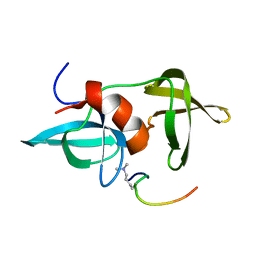 | |
2MWP
 
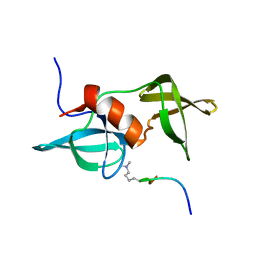 | |
2XWC
 
 | | Crystal structure of the DNA binding domain of human TP73 refined at 1.8 A resolution | | Descriptor: | GLYCEROL, TRIS(HYDROXYETHYL)AMINOMETHANE, TUMOUR PROTEIN P73, ... | | Authors: | Canning, P, Zhang, Y, Vollmar, M, Krojer, T, Ugochukwu, E, Muniz, J.R.C, von Delft, F, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Bullock, A.N. | | Deposit date: | 2010-11-03 | | Release date: | 2010-11-17 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Structural Basis for Aspp2 Recognition by the Tumor Suppressor P73.
J.Mol.Biol., 423, 2012
|
|
