8TJ5
 
 | | Inner spoke ring of the yeast NPC | | Descriptor: | Nucleoporin 59, Nucleoporin NIC96, Nucleoporin NSP1, ... | | Authors: | Akey, C.W, Echeverria, I, Ouch, C, Fernandez-Martinez, J, Rout, M.P. | | Deposit date: | 2023-07-20 | | Release date: | 2023-10-11 | | Method: | ELECTRON MICROSCOPY (6.6 Å) | | Cite: | Implications of a multiscale structure of the yeast nuclear pore complex.
Mol.Cell, 83, 2023
|
|
8TIE
 
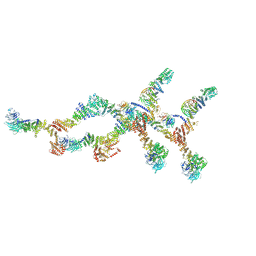 | | Double nuclear outer ring of Nup84-complexes from the yeast NPC | | Descriptor: | NUP133 isoform 1, NUP145 isoform 1, Nucleoporin NUP120, ... | | Authors: | Akey, C.W, Echeverria, I, Ouch, C, Fernandez-Martinez, J, Rout, M.P. | | Deposit date: | 2023-07-19 | | Release date: | 2023-10-11 | | Method: | ELECTRON MICROSCOPY (8.1 Å) | | Cite: | Implications of a multiscale structure of the yeast nuclear pore complex.
Mol.Cell, 83, 2023
|
|
8THU
 
 | |
8TAH
 
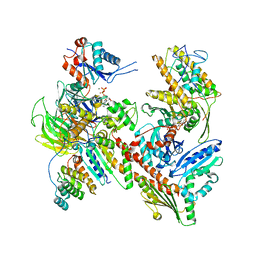 | | Cryo-EM structure of Cortactin-bound to Arp2/3 complex | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Actin-related protein 2, Actin-related protein 2/3 complex subunit 1A, ... | | Authors: | Fregoso, F.E, van Eeuwen, T, Dominguez, R. | | Deposit date: | 2023-06-27 | | Release date: | 2023-09-27 | | Last modified: | 2024-01-31 | | Method: | ELECTRON MICROSCOPY (2.89 Å) | | Cite: | Mechanism of synergistic activation of Arp2/3 complex by cortactin and WASP-family proteins.
Nat Commun, 14, 2023
|
|
8T9U
 
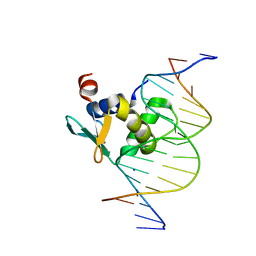 | | Human PU.1 ETS-domain (165-270) in complex with d(AATAAGCGIAAGTGGG) | | Descriptor: | DNA (5'-D(*AP*AP*TP*AP*AP*GP*CP*GP*IP*AP*AP*GP*TP*GP*GP*G)-3'), DNA (5'-D(*TP*CP*CP*CP*AP*CP*TP*TP*CP*CP*GP*CP*TP*TP*AP*T)-3'), SODIUM ION, ... | | Authors: | Terrell, J.R, Poon, G.M.K. | | Deposit date: | 2023-06-26 | | Release date: | 2023-11-29 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Dissection of integrated readout reveals the structural thermodynamics of DNA selection by transcription factors.
Structure, 32, 2024
|
|
8T9F
 
 | |
8T94
 
 | |
8T81
 
 | |
8T6Z
 
 | |
8T67
 
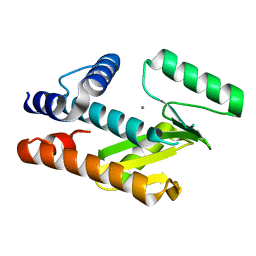 | |
8T5Z
 
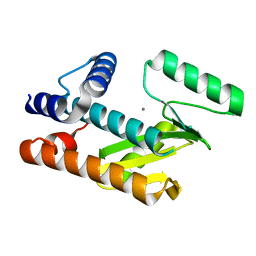 | |
8T5I
 
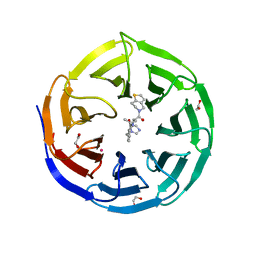 | | Crystal structure of human WDR5 in complex with MR4397 | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, N-[(2S)-1-(6,7-dihydrothieno[3,2-c]pyridin-5(4H)-yl)-1-oxopentan-2-yl]-3-[(1H-imidazol-1-yl)methyl]benzamide, ... | | Authors: | Kimani, S, Dong, A, Li, F, Loppnau, P, Ackloo, S, Vedadi, M, Brown, P.J, Arrowsmith, C.H, Edwards, A.M, Halabelian, L, Structural Genomics Consortium (SGC) | | Deposit date: | 2023-06-13 | | Release date: | 2023-08-09 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of human WDR5 in complex with MR4397
To be published
|
|
8T4S
 
 | |
8T2T
 
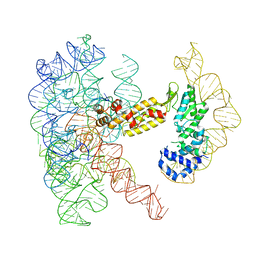 | | Structure of a group II intron ribonucleoprotein in the post-ligation (post-2F) state | | Descriptor: | AMMONIUM ION, Group II intron reverse transcriptase/maturase, MAGNESIUM ION, ... | | Authors: | Xu, L, Liu, T, Chung, K, Pyle, A.M. | | Deposit date: | 2023-06-06 | | Release date: | 2023-11-22 | | Last modified: | 2024-01-03 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural insights into intron catalysis and dynamics during splicing.
Nature, 624, 2023
|
|
8T2S
 
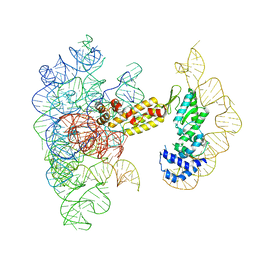 | | Structure of a group II intron ribonucleoprotein in the pre-branching (pre-1F) state | | Descriptor: | AMMONIUM ION, CALCIUM ION, Group II intron reverse transcriptase/maturase, ... | | Authors: | Xu, L, Liu, T, Chung, K, Pyle, A.M. | | Deposit date: | 2023-06-06 | | Release date: | 2023-11-22 | | Last modified: | 2024-01-03 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural insights into intron catalysis and dynamics during splicing.
Nature, 624, 2023
|
|
8T2R
 
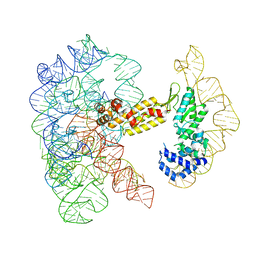 | | Structure of a group II intron ribonucleoprotein in the pre-ligation (pre-2F) state | | Descriptor: | 5'exon, AMMONIUM ION, CALCIUM ION, ... | | Authors: | Xu, L, Liu, T, Chung, K, Pyle, A.M. | | Deposit date: | 2023-06-06 | | Release date: | 2023-11-22 | | Last modified: | 2024-01-03 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural insights into intron catalysis and dynamics during splicing.
Nature, 624, 2023
|
|
8T1R
 
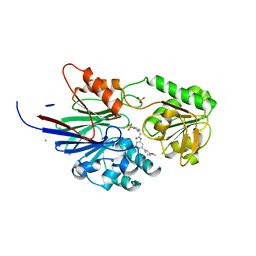 | | Crystal structure of human CPSF73 catalytic segment in complex with compound 2 | | Descriptor: | 3-[7,7-bis(oxidanyl)-8-oxa-7-boranuidabicyclo[4.3.0]nona-1,3,5-trien-5-yl]-~{N}-[3-(3-methoxyphenyl)phenyl]propanamide, CHLORIDE ION, Cleavage and polyadenylation specificity factor subunit 3, ... | | Authors: | Huang, J, Tong, L. | | Deposit date: | 2023-06-02 | | Release date: | 2023-11-08 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Anticancer benzoxaboroles block pre-mRNA processing by directly inhibiting CPSF3.
Cell Chem Biol, 31, 2024
|
|
8T1Q
 
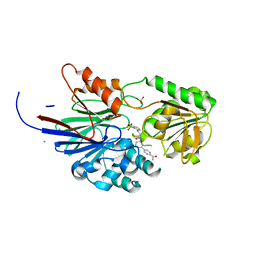 | | Crystal structure of human CPSF73 catalytic segment in complex with compound 1 | | Descriptor: | 3-[7,7-bis(oxidanyl)-8-oxa-7-boranuidabicyclo[4.3.0]nona-1,3,5-trien-5-yl]-~{N}-[3-(4-ethanoylphenyl)phenyl]propanamide, CHLORIDE ION, Cleavage and polyadenylation specificity factor subunit 3, ... | | Authors: | Huang, J, Tong, L. | | Deposit date: | 2023-06-02 | | Release date: | 2023-11-08 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Anticancer benzoxaboroles block pre-mRNA processing by directly inhibiting CPSF3.
Cell Chem Biol, 31, 2024
|
|
8T1K
 
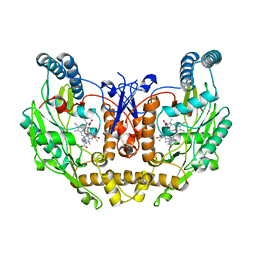 | | DSBU crosslinked nNOS-CaM oxygenase homodimer | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, ARGININE, Nitric oxide synthase 1, ... | | Authors: | Lee, K, Pospiech, T.H, Southworth, D. | | Deposit date: | 2023-06-02 | | Release date: | 2023-11-29 | | Last modified: | 2023-12-27 | | Method: | ELECTRON MICROSCOPY (3.14 Å) | | Cite: | Mapping interactions of calmodulin and neuronal NO synthase by crosslinking and mass spectrometry.
J.Biol.Chem., 300, 2023
|
|
8T1J
 
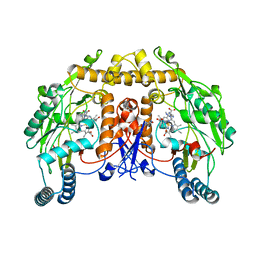 | | Uncrosslinked nNOS-CaM oxygenase homodimer | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, ARGININE, Nitric oxide synthase 1, ... | | Authors: | Lee, K, Pospiech, T.H, Southworth, D. | | Deposit date: | 2023-06-02 | | Release date: | 2023-11-29 | | Last modified: | 2023-12-27 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Mapping interactions of calmodulin and neuronal NO synthase by crosslinking and mass spectrometry.
J.Biol.Chem., 300, 2023
|
|
8T1F
 
 | | Cryo-EM structure of full-length human TRPV4 in complex with antagonist HC-067047 | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, 2-methyl-1-[3-(morpholin-4-yl)propyl]-5-phenyl-N-[3-(trifluoromethyl)phenyl]-1H-pyrrole-3-carboxamide, Transient receptor potential cation channel subfamily V member 4/Enhanced green fluorescent protein chimera | | Authors: | Talyzina, I.A, Nadezhdin, K.D, Neuberger, A, Sobolevsky, A.I. | | Deposit date: | 2023-06-02 | | Release date: | 2023-07-05 | | Method: | ELECTRON MICROSCOPY (3.49 Å) | | Cite: | Structure of human TRPV4 in complex with GTPase RhoA.
Nat Commun, 14, 2023
|
|
8T1E
 
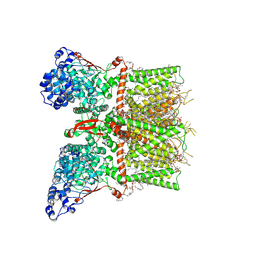 | | Closed-state cryo-EM structure of full-length human TRPV4 in the presence of 4a-PDD | | Descriptor: | (2R)-2-{[(4-O-hexopyranosyl-beta-D-glucopyranosyl)oxy]methyl}-4-{[(25R)-5beta,14beta,17beta-spirostan-3beta-yl]oxy}butyl 4-O-alpha-D-glucopyranosyl-beta-D-glucopyranoside, SODIUM ION, Transient receptor potential cation channel subfamily V member 4,Enhanced green fluorescent protein, ... | | Authors: | Talyzina, I.A, Nadezhdin, K.D, Neuberger, A, Sobolevsky, A.I. | | Deposit date: | 2023-06-02 | | Release date: | 2023-07-05 | | Method: | ELECTRON MICROSCOPY (2.77 Å) | | Cite: | Structure of human TRPV4 in complex with GTPase RhoA.
Nat Commun, 14, 2023
|
|
8T1D
 
 | | Open-state cryo-EM structure of full-length human TRPV4 in complex with agonist 4a-PDD | | Descriptor: | (1aR,1bS,4aS,7aS,7bS,8R,9R,9aS)-9a-(decanoyloxy)-4a,7b-dihydroxy-3-(hydroxymethyl)-1,1,6,8-tetramethyl-5-oxo-1a,1b,4,4a,5,7a,7b,8,9,9a-decahydro-1H-cyclopropa[3,4]benzo[1,2-e]azulen-9-yl decanoate, Transient receptor potential cation channel subfamily V member 4/Enhanced green fluorescent protein chimera | | Authors: | Talyzina, I.A, Nadezhdin, K.D, Neuberger, A, Sobolevsky, A.I. | | Deposit date: | 2023-06-02 | | Release date: | 2023-07-05 | | Method: | ELECTRON MICROSCOPY (3.35 Å) | | Cite: | Structure of human TRPV4 in complex with GTPase RhoA.
Nat Commun, 14, 2023
|
|
8T1C
 
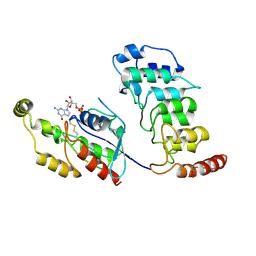 | | Cryo-EM structure of human TRPV4 ankyrin repeat domain in complex with GTPase RhoA | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, Transforming protein RhoA, Transient receptor potential cation channel subfamily V member 4-Enhanced green fluorescent protein chimera | | Authors: | Nadezhdin, K.D, Talyzina, I.A, Neuberger, A, Sobolevsky, A.I. | | Deposit date: | 2023-06-02 | | Release date: | 2023-07-05 | | Method: | ELECTRON MICROSCOPY (3.49 Å) | | Cite: | Structure of human TRPV4 in complex with GTPase RhoA.
Nat Commun, 14, 2023
|
|
8T1B
 
 | | Cryo-EM structure of full-length human TRPV4 in apo state | | Descriptor: | (2R)-2-{[(4-O-hexopyranosyl-beta-D-glucopyranosyl)oxy]methyl}-4-{[(25R)-5beta,14beta,17beta-spirostan-3beta-yl]oxy}butyl 4-O-alpha-D-glucopyranosyl-beta-D-glucopyranoside, SODIUM ION, Transient receptor potential cation channel subfamily V member 4/Enhanced green fluorescent protein chimera, ... | | Authors: | Nadezhdin, K.D, Talyzina, I.A, Neuberger, A, Sobolevsky, A.I. | | Deposit date: | 2023-06-02 | | Release date: | 2023-07-05 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structure of human TRPV4 in complex with GTPase RhoA.
Nat Commun, 14, 2023
|
|
