4YJE
 
 | | Crystal structure of APC-ARM in complexed with Amer1-A1 | | Descriptor: | APC membrane recruitment protein 1, Adenomatous polyposis coli protein | | Authors: | Zhang, Z, Xiao, Y, Wu, G. | | Deposit date: | 2015-03-03 | | Release date: | 2016-03-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structures of the APC-ARM domain in complexes with discrete Amer1/WTX fragments reveal that it uses a consensus mode to recognize its binding partners
Cell Discov, 1, 2015
|
|
4YJL
 
 | | Crystal structure of APC-ARM in complexed with Amer1-A2 | | Descriptor: | 1,2-ETHANEDIOL, APC membrane recruitment protein 1, Adenomatous polyposis coli protein | | Authors: | Zhang, Z, Xiao, Y, Wu, G. | | Deposit date: | 2015-03-03 | | Release date: | 2016-03-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structures of the APC-ARM domain in complexes with discrete Amer1/WTX fragments reveal that it uses a consensus mode to recognize its binding partners
Cell Discov, 1, 2015
|
|
4YI0
 
 | | Structure of mouse importin a1 bound to Pom121NLS | | Descriptor: | Importin subunit alpha-1, Nuclear envelope pore membrane protein POM 121 | | Authors: | Lokareddy, R.K, Cingolani, G. | | Deposit date: | 2015-02-27 | | Release date: | 2016-01-13 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.806 Å) | | Cite: | Mammalian Pom121 and yeast Heh2 share IBB-like NLSs that support targeting to the inner nuclear membrane
To Be Published
|
|
1EE4
 
 | |
1EJY
 
 | |
1EJL
 
 | |
4YK6
 
 | | Crystal structure of APC-ARM in complexed with Amer1-A4 | | Descriptor: | APC membrane recruitment protein 1, Adenomatous polyposis coli protein | | Authors: | Zhang, Z, Xiao, Y, Wu, G. | | Deposit date: | 2015-03-04 | | Release date: | 2016-03-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structures of the APC-ARM domain in complexes with discrete Amer1/WTX fragments reveal that it uses a consensus mode to recognize its binding partners
Cell Discov, 1, 2015
|
|
1EE5
 
 | |
4ZDU
 
 | |
6WBB
 
 | | Structure of Mouse Importin alpha - MLH1-E475A NLS peptide complex | | Descriptor: | DNA mismatch repair protein Mlh1, Importin subunit alpha-1 | | Authors: | De Barros, A.C, Da Silva, T.D, Oliveira, H.C, Fukuda, C.A, Fontes, M.R.M. | | Deposit date: | 2020-03-26 | | Release date: | 2021-07-07 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.663 Å) | | Cite: | Structural and calorimetric studies reveal specific determinants for the binding of a high-affinity NLS to mammalian importin-alpha.
Biochem.J., 478, 2021
|
|
6WBA
 
 | | Structure of Mouse Importin alpha MLH1- R470A NLS Peptide Complex | | Descriptor: | DNA mismatch repair protein Mlh1, Importin subunit alpha-1 | | Authors: | de Barros, A.C, da Silva, T.D, Oliveira, H.C, Fukuda, C.A, Fontes, M.R.M. | | Deposit date: | 2020-03-26 | | Release date: | 2021-07-07 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.151 Å) | | Cite: | Structural and calorimetric studies reveal specific determinants for the binding of a high-affinity NLS to mammalian importin-alpha.
Biochem.J., 478, 2021
|
|
6WBC
 
 | | Structure of Mouse Importin alpha- MLH1-R472K NLS Peptide Complex | | Descriptor: | DNA mismatch repair protein Mlh1, Importin subunit alpha-1 | | Authors: | de Barros, A.C, da Silva, T.D, Oliveira, H.C, Fukuda, C.A, Fontes, M.R.M. | | Deposit date: | 2020-03-26 | | Release date: | 2021-07-07 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural and calorimetric studies reveal specific determinants for the binding of a high-affinity NLS to mammalian importin-alpha.
Biochem.J., 478, 2021
|
|
1F59
 
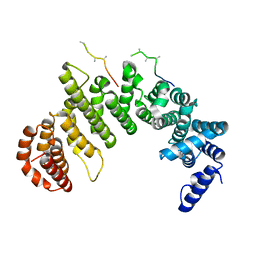 | | IMPORTIN-BETA-FXFG NUCLEOPORIN COMPLEX | | Descriptor: | FXFG NUCLEOPORIN REPEATS, IMPORTIN BETA-1 | | Authors: | Bayliss, R, Littlewood, T, Stewart, M. | | Deposit date: | 2000-06-13 | | Release date: | 2000-08-16 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis for the interaction between FxFG nucleoporin repeats and importin-beta in nuclear trafficking.
Cell(Cambridge,Mass.), 102, 2000
|
|
1T08
 
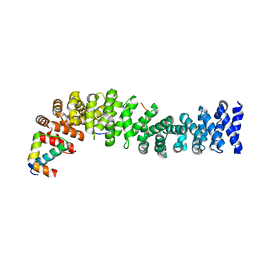 | | Crystal structure of beta-catenin/ICAT helical domain/unphosphorylated APC R3 | | Descriptor: | Adenomatous polyposis coli protein, Beta-catenin, Beta-catenin-interacting protein 1 | | Authors: | Ha, N.-C, Tonozuka, T, Stamos, J.L, Weis, W.I. | | Deposit date: | 2004-04-07 | | Release date: | 2004-10-12 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Mechanism of phosphorylation-dependent binding of APC to beta-catenin and its role in beta-catenin degradation
Mol.Cell, 15, 2004
|
|
1TH1
 
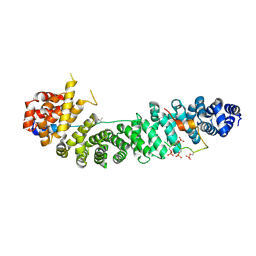 | | Beta-catenin in complex with a phosphorylated APC 20aa repeat fragment | | Descriptor: | Adenomatous polyposis coli protein, Beta-catenin | | Authors: | Xing, Y, Clements, W.K, Le Trong, I, Hinds, T.R, Stenkamp, R, Kimelman, D, Xu, W. | | Deposit date: | 2004-05-31 | | Release date: | 2004-09-07 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of a beta-Catenin/APC Complex Reveals a Critical Role for APC Phosphorylation in APC Function.
Mol.Cell, 15, 2004
|
|
1V18
 
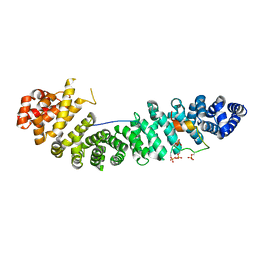 | |
1UN0
 
 | | Crystal Structure of Yeast Karyopherin (Importin) alpha in complex with a Nup2p N-terminal fragment | | Descriptor: | IMPORTIN ALPHA SUBUNIT, NUCLEOPORIN NUP2 | | Authors: | Matsuura, Y, Lange, A, Harreman, M.T, Corbett, A.H, Stewart, M. | | Deposit date: | 2003-09-03 | | Release date: | 2003-10-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural Basis for Nup2P Function in Cargo Release and Karyopherin Recycling in Nuclear Import
Embo J., 22, 2003
|
|
1Y2A
 
 | | Structure of mammalian importin bound to the non-classical PLSCR1-NLS | | Descriptor: | Importin alpha-2 Subunit, decamer fragment of Phospholipid scramblase 1 | | Authors: | Chen, M.-H, Ben-Efraim, I, Mitrousis, G, Walker-Kopp, N, Sims, P.J, Cingolani, G. | | Deposit date: | 2004-11-22 | | Release date: | 2005-02-01 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Phospholipid Scramblase 1 Contains a Nonclassical Nuclear Localization Signal with Unique Binding Site in Importin alpha
J.Biol.Chem., 280, 2005
|
|
5B56
 
 | | Crystal structure of HIV-1 VPR C-Terminal domain and DIBB-M-Importin-Alpha2 complex | | Descriptor: | Importin subunit alpha-1, Protein Vpr | | Authors: | Miyatake, H, Sanjoh, A, Matusda, G, Murakami, T, Murakami, H, Hagiwara, K, Yokoyama, M, Sato, H, Miyamoto, Y, Dohmae, N, Aida, Y. | | Deposit date: | 2016-04-25 | | Release date: | 2016-06-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Molecular Mechanism of HIV-1 Vpr for Binding to Importin-alpha
J.Mol.Biol., 428, 2016
|
|
5B6G
 
 | | Protein-protein interaction | | Descriptor: | Adenomatous polyposis coli protein, GLYCEROL, PHQ-ALA-GLY-GLU-ALA-XYC-TYR-GLU, ... | | Authors: | Zhao, Y, Jiang, H, Yang, X, Jiang, F, Song, K, Zhang, J. | | Deposit date: | 2016-05-27 | | Release date: | 2017-05-31 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Peptidomimetic inhibitors of APC-Asef interaction block colorectal cancer migration.
Nat. Chem. Biol., 13, 2017
|
|
7TMX
 
 | |
7TMY
 
 | |
5CTT
 
 | |
5D5K
 
 | |
7UMI
 
 | | Importin a1 bound to Cp183-CTD | | Descriptor: | HBV-NLS, Importin subunit alpha-1 | | Authors: | Yang, R, Cingolani, G. | | Deposit date: | 2022-04-07 | | Release date: | 2023-10-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Structural basis for nuclear import of hepatitis B virus (HBV) nucleocapsid core.
Sci Adv, 10, 2024
|
|
