4MV3
 
 | | Crystal Structure of Biotin Carboxylase from Haemophilus influenzae in Complex with AMPPCP and Bicarbonate | | Descriptor: | 1,2-ETHANEDIOL, BICARBONATE ION, Biotin carboxylase, ... | | Authors: | Broussard, T.C, Pakhomova, S, Neau, D.B, Champion, T.S, Bonnot, R.J, Waldrop, G.L. | | Deposit date: | 2013-09-23 | | Release date: | 2015-01-14 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Structural Analysis of Substrate, Reaction Intermediate, and Product Binding in Haemophilus influenzae Biotin Carboxylase.
Biochemistry, 54, 2015
|
|
4MV7
 
 | | Crystal Structure of Biotin Carboxylase form Haemophilus influenzae in Complex with Phosphonoformate | | Descriptor: | 1,2-ETHANEDIOL, Biotin carboxylase, PHOSPHONOFORMIC ACID | | Authors: | Broussard, T.C, Pakhomova, S, Neau, D.B, Champion, T.S, Bonnot, R.J, Waldrop, G.L. | | Deposit date: | 2013-09-23 | | Release date: | 2015-01-14 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Structural Analysis of Substrate, Reaction Intermediate, and Product Binding in Haemophilus influenzae Biotin Carboxylase.
Biochemistry, 54, 2015
|
|
4MV6
 
 | | Crystal Structure of Biotin Carboxylase from Haemophilus influenzae in Complex with Phosphonoacetamide | | Descriptor: | 1,2-ETHANEDIOL, Biotin carboxylase, PHOSPHONOACETAMIDE | | Authors: | Broussard, T.C, Pakhomova, S, Neau, D.B, Champion, T.S, Bonnot, R.J, Waldrop, G.L. | | Deposit date: | 2013-09-23 | | Release date: | 2015-01-14 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Structural Analysis of Substrate, Reaction Intermediate, and Product Binding in Haemophilus influenzae Biotin Carboxylase.
Biochemistry, 54, 2015
|
|
3GLK
 
 | |
3HB9
 
 | | Crystal Structure of S. aureus Pyruvate Carboxylase A610T Mutant | | Descriptor: | 5-(HEXAHYDRO-2-OXO-1H-THIENO[3,4-D]IMIDAZOL-6-YL)PENTANAL, ADENOSINE-5'-DIPHOSPHATE, MANGANESE (II) ION, ... | | Authors: | Tong, L, Yu, L.P.C. | | Deposit date: | 2009-05-04 | | Release date: | 2009-06-30 | | Last modified: | 2021-10-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | A Symmetrical Tetramer for S. aureus Pyruvate Carboxylase in Complex with Coenzyme A.
Structure, 17, 2009
|
|
3JZF
 
 | |
3JZI
 
 | | Crystal structure of biotin carboxylase from E. Coli in complex with benzimidazole series | | Descriptor: | 7-amino-2-[(2-chlorobenzyl)amino]-1-{[(1S,2S)-2-hydroxycycloheptyl]methyl}-1H-benzimidazole-5-carboxamide, Biotin carboxylase | | Authors: | Cheng, C, Shipps, G.W, Yang, Z, Sun, B, Kawahata, N, Soucy, K, Soriano, A, Orth, P, Xiao, L, Mann, P, Black, T. | | Deposit date: | 2009-09-23 | | Release date: | 2009-11-03 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Discovery and optimization of antibacterial AccC inhibitors.
Bioorg.Med.Chem.Lett., 19, 2009
|
|
3JRW
 
 | | Phosphorylated BC domain of ACC2 | | Descriptor: | Acetyl-CoA carboxylase 2 | | Authors: | Cho, Y.S, Lee, J.I, Shin, D, Kim, H.T, Lee, T.G, Heo, Y.S. | | Deposit date: | 2009-09-09 | | Release date: | 2010-01-12 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Molecular mechanism for the regulation of human ACC2 through phosphorylation by AMPK
Biochem.Biophys.Res.Commun., 391, 2010
|
|
3HO8
 
 | |
3HBL
 
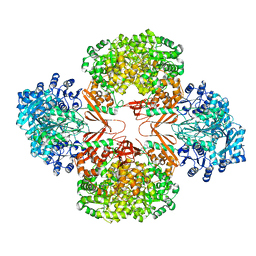 | | Crystal Structure of S. aureus Pyruvate Carboxylase T908A Mutant | | Descriptor: | 5-(HEXAHYDRO-2-OXO-1H-THIENO[3,4-D]IMIDAZOL-6-YL)PENTANAL, ADENOSINE-5'-DIPHOSPHATE, MANGANESE (II) ION, ... | | Authors: | Tong, L, Yu, L.P.C. | | Deposit date: | 2009-05-04 | | Release date: | 2009-06-30 | | Last modified: | 2021-10-13 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | A Symmetrical Tetramer for S. aureus Pyruvate Carboxylase in Complex with Coenzyme A.
Structure, 17, 2009
|
|
3JRX
 
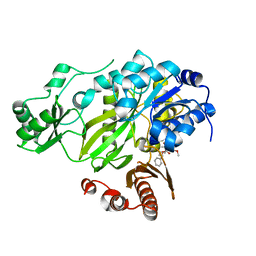 | | Crystal structure of the BC domain of ACC2 in complex with soraphen A | | Descriptor: | Acetyl-CoA carboxylase 2, SORAPHEN A | | Authors: | Cho, Y.S, Lee, J.I, Shin, D, Kim, H.T, Lee, T.G, Heo, Y.S. | | Deposit date: | 2009-09-09 | | Release date: | 2010-01-12 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Molecular mechanism for the regulation of human ACC2 through phosphorylation by AMPK.
Biochem.Biophys.Res.Commun., 391, 2010
|
|
7YBU
 
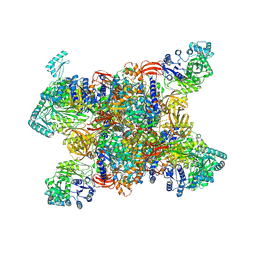 | | Human propionyl-coenzyme A carboxylase | | Descriptor: | 5-(HEXAHYDRO-2-OXO-1H-THIENO[3,4-D]IMIDAZOL-6-YL)PENTANAL, Propionyl-CoA carboxylase alpha chain, mitochondrial, ... | | Authors: | Su, J.Y, Liu, D.S. | | Deposit date: | 2022-06-29 | | Release date: | 2023-07-05 | | Method: | ELECTRON MICROSCOPY (2.2 Å) | | Cite: | Human propionyl-coenzyme A carboxylase
To Be Published
|
|
7ZZ5
 
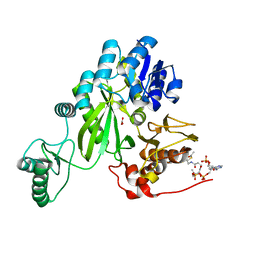 | | Cryo-EM structure of "BC open" conformation of Lactococcus lactis pyruvate carboxylase with acetyl-CoA | | Descriptor: | ACETYL COENZYME *A, BICARBONATE ION, Pyruvate carboxylase | | Authors: | Lopez-Alonso, J.P, Lazaro, M, Gil, D, Choi, P.H, Tong, L, Valle, M. | | Deposit date: | 2022-05-25 | | Release date: | 2022-10-12 | | Last modified: | 2022-10-26 | | Method: | ELECTRON MICROSCOPY (2.43 Å) | | Cite: | CryoEM structural exploration of catalytically active enzyme pyruvate carboxylase.
Nat Commun, 13, 2022
|
|
7ZZ3
 
 | | Cryo-EM structure of "BC react" conformation of Lactococcus lactis pyruvate carboxylase with acetyl-CoA | | Descriptor: | ACETYL COENZYME *A, ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Lopez-Alonso, J.P, Lazaro, M, Gil, D, Choi, P.H, Tong, L, Valle, M. | | Deposit date: | 2022-05-25 | | Release date: | 2022-10-12 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (2.41 Å) | | Cite: | CryoEM structural exploration of catalytically active enzyme pyruvate carboxylase.
Nat Commun, 13, 2022
|
|
7ZYZ
 
 | | Cryo-EM structure of "CT oxa" conformation of Lactococcus lactis pyruvate carboxylase with acetyl-CoA | | Descriptor: | MANGANESE (II) ION, OXALOACETATE ION, Pyruvate carboxylase | | Authors: | Lopez-Alonso, J.P, Lazaro, M, Gil, D, Choi, P.H, Tong, L, Valle, M. | | Deposit date: | 2022-05-25 | | Release date: | 2022-10-12 | | Last modified: | 2022-11-02 | | Method: | ELECTRON MICROSCOPY (2.47 Å) | | Cite: | CryoEM structural exploration of catalytically active enzyme pyruvate carboxylase.
Nat Commun, 13, 2022
|
|
7ZZ6
 
 | | Cryo-EM structure of "CT-CT dimer" of Lactococcus lactis pyruvate carboxylase with acetyl-CoA | | Descriptor: | MAGNESIUM ION, MANGANESE (II) ION, PYRUVIC ACID, ... | | Authors: | Lopez-Alonso, J.P, Lazaro, M, Gil, D, Choi, P.H, Tong, L, Valle, M. | | Deposit date: | 2022-05-25 | | Release date: | 2022-12-28 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (2.15 Å) | | Cite: | CryoEM structural exploration of catalytically active enzyme pyruvate carboxylase.
Nat Commun, 13, 2022
|
|
7ZZ8
 
 | | Cryo-EM structure of Lactococcus lactis pyruvate carboxylase with acetyl-CoA and cyclic di-AMP | | Descriptor: | (2R,3R,3aS,5R,7aR,9R,10R,10aS,12R,14aR)-2,9-bis(6-amino-9H-purin-9-yl)octahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8 ]tetraoxadiphosphacyclododecine-3,5,10,12-tetrol 5,12-dioxide, ACETYL COENZYME *A, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Lopez-Alonso, J.P, Lazaro, M, Gil, D, Choi, P.H, Tong, L, Valle, M. | | Deposit date: | 2022-05-25 | | Release date: | 2022-10-12 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (3.29 Å) | | Cite: | CryoEM structural exploration of catalytically active enzyme pyruvate carboxylase.
Nat Commun, 13, 2022
|
|
7ZZ1
 
 | | Cryo-EM structure of "CT react" conformation of Lactococcus lactis pyruvate carboxylase with acetyl-CoA | | Descriptor: | BIOTIN, MAGNESIUM ION, MANGANESE (II) ION, ... | | Authors: | Lopez-Alonso, J.P, Lazaro, M, Gil, D, Choi, P.H, Tong, L, Valle, M. | | Deposit date: | 2022-05-25 | | Release date: | 2022-10-12 | | Last modified: | 2022-11-02 | | Method: | ELECTRON MICROSCOPY (2.27 Å) | | Cite: | CryoEM structural exploration of catalytically active enzyme pyruvate carboxylase.
Nat Commun, 13, 2022
|
|
7ZZ2
 
 | | Cryo-EM structure of "CT pyr" conformation of Lactococcus lactis pyruvate carboxylase with acetyl-CoA | | Descriptor: | MAGNESIUM ION, MANGANESE (II) ION, PYRUVIC ACID, ... | | Authors: | Lopez-Alonso, J.P, Lazaro, M, Gil, D, Choi, P.H, Tong, L, Valle, M. | | Deposit date: | 2022-05-25 | | Release date: | 2022-10-12 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (2.48 Å) | | Cite: | CryoEM structural exploration of catalytically active enzyme pyruvate carboxylase.
Nat Commun, 13, 2022
|
|
7ZZ0
 
 | | Cryo-EM structure of "CT empty" conformation of Lactococcus lactis pyruvate carboxylase with acetyl-CoA | | Descriptor: | MAGNESIUM ION, MANGANESE (II) ION, Pyruvate carboxylase | | Authors: | Lopez-Alonso, J.P, Lazaro, M, Gil, D, Choi, P.H, Tong, L, Valle, M. | | Deposit date: | 2022-05-25 | | Release date: | 2022-10-12 | | Last modified: | 2022-11-02 | | Method: | ELECTRON MICROSCOPY (2.26 Å) | | Cite: | CryoEM structural exploration of catalytically active enzyme pyruvate carboxylase.
Nat Commun, 13, 2022
|
|
7ZZ4
 
 | | Cryo-EM structure of "BC closed" conformation of Lactococcus lactis pyruvate carboxylase with acetyl-CoA | | Descriptor: | ACETYL COENZYME *A, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Lopez-Alonso, J.P, Lazaro, M, Gil, D, Choi, P.H, Tong, L, Valle, M. | | Deposit date: | 2022-05-25 | | Release date: | 2022-10-12 | | Last modified: | 2022-11-02 | | Method: | ELECTRON MICROSCOPY (2.63 Å) | | Cite: | CryoEM structural exploration of catalytically active enzyme pyruvate carboxylase.
Nat Commun, 13, 2022
|
|
7ZYY
 
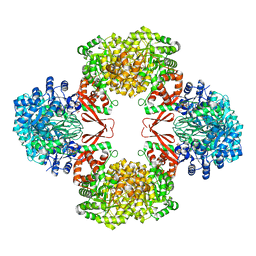 | | Cryo-EM structure of Lactococcus lactis pyruvate carboxylase with acetyl-CoA | | Descriptor: | ACETYL COENZYME *A, ADENOSINE-5'-DIPHOSPHATE, BICARBONATE ION, ... | | Authors: | Lopez-Alonso, J.P, Lazaro, M, Gil, D, Choi, P.H, Tong, L, Valle, M. | | Deposit date: | 2022-05-25 | | Release date: | 2022-10-12 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (2.12 Å) | | Cite: | CryoEM structural exploration of catalytically active enzyme pyruvate carboxylase.
Nat Commun, 13, 2022
|
|
2QF7
 
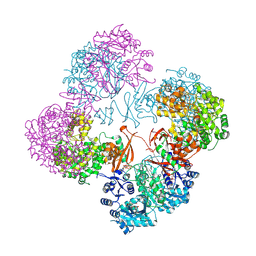 | | Crystal structure of a complete multifunctional pyruvate carboxylase from Rhizobium etli | | Descriptor: | CHLORIDE ION, COENZYME A, FORMIC ACID, ... | | Authors: | St Maurice, M, Surinya, K.H, Rayment, I. | | Deposit date: | 2007-06-27 | | Release date: | 2007-09-04 | | Last modified: | 2012-02-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Domain architecture of pyruvate carboxylase, a biotin-dependent multifunctional enzyme
Science, 317, 2007
|
|
2V59
 
 | |
2W6Z
 
 | |
