4C02
 
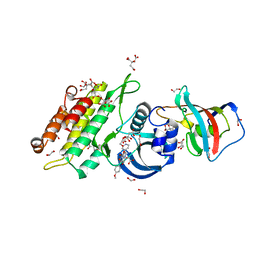 | | Crystal structure of human ACVR1 (ALK2) in complex with FKBP12.6 and dorsomorphin | | Descriptor: | 1,2-ETHANEDIOL, 6-[4-(2-piperidin-1-ylethoxy)phenyl]-3-pyridin-4-ylpyrazolo[1,5-a]pyrimidine, ACTIVIN RECEPTOR TYPE-1, ... | | Authors: | Williams, E, Riesebos, E, Vollmar, M, Krojer, T, Bradley, A, Shrestha, L, Kupinska, K, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Bullock, A. | | Deposit date: | 2013-07-31 | | Release date: | 2013-08-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Crystal Structure of Human Acvr1 (Alk2) in Complex with Fkbp12.6 And Dorsomorphin
Ph D Thesis
|
|
4DRH
 
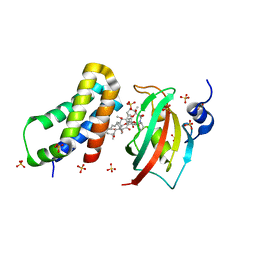 | | Co-crystal structure of the PPIase domain of FKBP51, Rapamycin and the FRB fragment of mTOR at low pH | | Descriptor: | Peptidyl-prolyl cis-trans isomerase FKBP5, RAPAMYCIN IMMUNOSUPPRESSANT DRUG, SULFATE ION, ... | | Authors: | Maerz, A.M, Bracher, A, Hausch, F. | | Deposit date: | 2012-02-17 | | Release date: | 2013-02-06 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Large FK506-Binding Proteins Shape the Pharmacology of Rapamycin.
Mol.Cell.Biol., 33, 2013
|
|
3MDY
 
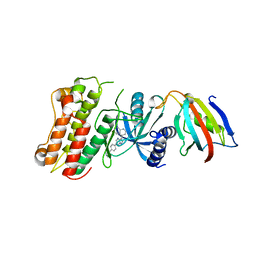 | | Crystal structure of the cytoplasmic domain of the bone morphogenetic protein receptor type-1B (BMPR1B) in complex with FKBP12 and LDN-193189 | | Descriptor: | 4-[6-(4-piperazin-1-ylphenyl)pyrazolo[1,5-a]pyrimidin-3-yl]quinoline, Bone morphogenetic protein receptor type-1B, Peptidyl-prolyl cis-trans isomerase FKBP1A | | Authors: | Chaikuad, A, Sanvitale, C, Mahajan, P, Daga, N, Cooper, C, Krojer, T, Alfano, I, Knapp, S, von Delft, F, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Bullock, A, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-03-31 | | Release date: | 2010-05-19 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal structure of the cytoplasmic domain of the bone morphogenetic protein receptor type-1B (BMPR1B) in complex with FKBP12 and LDN-193189
To be Published
|
|
4DRI
 
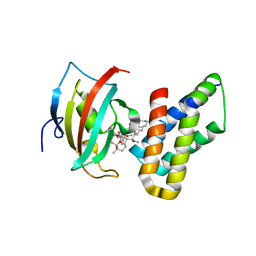 | | Co-crystal structure of the PPIase domain of FKBP51, Rapamycin and the FRB fragment of mTOR | | Descriptor: | Peptidyl-prolyl cis-trans isomerase FKBP5, RAPAMYCIN IMMUNOSUPPRESSANT DRUG, Serine/threonine-protein kinase mTOR | | Authors: | Maerz, A.M, Bracher, A, Hausch, F. | | Deposit date: | 2012-02-17 | | Release date: | 2013-02-06 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Large FK506-Binding Proteins Shape the Pharmacology of Rapamycin.
Mol.Cell.Biol., 33, 2013
|
|
4DRJ
 
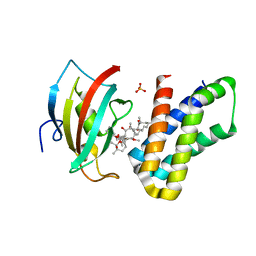 | | o-crystal structure of the PPIase domain of FKBP52, Rapamycin and the FRB fragment of mTOR | | Descriptor: | Peptidyl-prolyl cis-trans isomerase FKBP4, RAPAMYCIN IMMUNOSUPPRESSANT DRUG, SULFATE ION, ... | | Authors: | Maerz, A.M, Bracher, A, Hausch, F. | | Deposit date: | 2012-02-17 | | Release date: | 2013-02-06 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Large FK506-Binding Proteins Shape the Pharmacology of Rapamycin.
Mol.Cell.Biol., 33, 2013
|
|
8GL6
 
 | |
8GLJ
 
 | |
8GLK
 
 | |
8GLN
 
 | |
8GLM
 
 | |
8GL8
 
 | |
1FD9
 
 | | CRYSTAL STRUCTURE OF THE MACROPHAGE INFECTIVITY POTENTIATOR PROTEIN (MIP) A MAJOR VIRULENCE FACTOR FROM LEGIONELLA PNEUMOPHILA | | Descriptor: | PROTEIN (MACROPHAGE INFECTIVITY POTENTIATOR PROTEIN), ZINC ION | | Authors: | Riboldi-Tunnicliffe, A, Jessen, S, Konig, B, Rahfeld, J, Hacker, J, Fischer, G, Hilgenfeld, R. | | Deposit date: | 2000-07-20 | | Release date: | 2001-07-25 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Crystal structure of Mip, a prolylisomerase from Legionella pneumophila
Nat.Struct.Biol., 8, 2001
|
|
1FAP
 
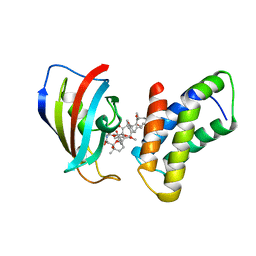 | | THE STRUCTURE OF THE IMMUNOPHILIN-IMMUNOSUPPRESSANT FKBP12-RAPAMYCIN COMPLEX INTERACTING WITH HUMAN FRAP | | Descriptor: | FK506-BINDING PROTEIN, FRAP, RAPAMYCIN IMMUNOSUPPRESSANT DRUG | | Authors: | Choi, J, Chen, J, Schreiber, S.L, Clardy, J. | | Deposit date: | 1996-03-15 | | Release date: | 1997-07-23 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure of the FKBP12-rapamycin complex interacting with the binding domain of human FRAP.
Science, 273, 1996
|
|
7VML
 
 | | Structure of recombinant RyR2 (EGTA dataset, class 1&2, closed state) | | Descriptor: | Peptidyl-prolyl cis-trans isomerase FKBP1B, Ryanodine receptor 2, ZINC ION | | Authors: | Kobayashi, T, Tsutsumi, A, Kurebayashi, N, Kodama, M, Kikkawa, M, Murayama, T, Ogawa, H. | | Deposit date: | 2021-10-09 | | Release date: | 2022-08-10 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Molecular basis for gating of cardiac ryanodine receptor explains the mechanisms for gain- and loss-of function mutations.
Nat Commun, 13, 2022
|
|
7VMN
 
 | | Structure of recombinant RyR2 (EGTA dataset, class 2, closed state) | | Descriptor: | Peptidyl-prolyl cis-trans isomerase FKBP1B, Ryanodine receptor 2, ZINC ION | | Authors: | Kobayashi, T, Tsutsumi, A, Kurebayashi, N, Kodama, M, Kikkawa, M, Murayama, T, Ogawa, H. | | Deposit date: | 2021-10-09 | | Release date: | 2022-08-10 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Molecular basis for gating of cardiac ryanodine receptor explains the mechanisms for gain- and loss-of function mutations.
Nat Commun, 13, 2022
|
|
7VMO
 
 | | Structure of recombinant RyR2 (Ca2+ dataset, class 1, open state) | | Descriptor: | CALCIUM ION, Peptidyl-prolyl cis-trans isomerase FKBP1B, Ryanodine receptor 2, ... | | Authors: | Kobayashi, T, Tsutsumi, A, Kurebayashi, N, Kodama, M, Kikkawa, M, Murayama, T, Ogawa, H. | | Deposit date: | 2021-10-09 | | Release date: | 2022-08-10 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Molecular basis for gating of cardiac ryanodine receptor explains the mechanisms for gain- and loss-of function mutations.
Nat Commun, 13, 2022
|
|
7VMP
 
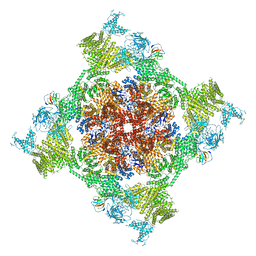 | | Structure of recombinant RyR2 (Ca2+ dataset, class 2, open state) | | Descriptor: | CALCIUM ION, Peptidyl-prolyl cis-trans isomerase FKBP1B, Ryanodine receptor 2, ... | | Authors: | Kobayashi, T, Tsutsumi, A, Kurebayashi, N, Kodama, M, Kikkawa, M, Murayama, T, Ogawa, H. | | Deposit date: | 2021-10-09 | | Release date: | 2022-08-10 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Molecular basis for gating of cardiac ryanodine receptor explains the mechanisms for gain- and loss-of function mutations.
Nat Commun, 13, 2022
|
|
7VMQ
 
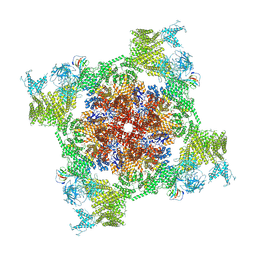 | | Structure of recombinant RyR2 (Ca2+ dataset, class 3, open state) | | Descriptor: | CALCIUM ION, Peptidyl-prolyl cis-trans isomerase FKBP1B, Ryanodine receptor 2, ... | | Authors: | Kobayashi, T, Tsutsumi, A, Kurebayashi, N, Kodama, M, Kikkawa, M, Murayama, T, Ogawa, H. | | Deposit date: | 2021-10-09 | | Release date: | 2022-08-10 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Molecular basis for gating of cardiac ryanodine receptor explains the mechanisms for gain- and loss-of function mutations
Nat Commun, 13, 2022
|
|
7VMM
 
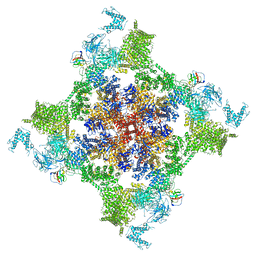 | | Structure of recombinant RyR2 (EGTA dataset, class 1, closed state) | | Descriptor: | Peptidyl-prolyl cis-trans isomerase FKBP1B, Ryanodine receptor 2, ZINC ION | | Authors: | Kobayashi, T, Tsutsumi, A, Kurebayashi, N, Kodama, M, Kikkawa, M, Murayama, T, Ogawa, H. | | Deposit date: | 2021-10-09 | | Release date: | 2022-08-10 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Molecular basis for gating of cardiac ryanodine receptor explains the mechanisms for gain- and loss-of function mutations.
Nat Commun, 13, 2022
|
|
7VMS
 
 | | Structure of recombinant RyR2 mutant K4593A (Ca2+ dataset) | | Descriptor: | CALCIUM ION, Peptidyl-prolyl cis-trans isomerase FKBP1B, Ryanodine receptor 2, ... | | Authors: | Kobayashi, T, Tsutsumi, A, Kurebayashi, N, Kodama, M, Kikkawa, M, Murayama, T, Ogawa, H. | | Deposit date: | 2021-10-09 | | Release date: | 2022-08-10 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Molecular basis for gating of cardiac ryanodine receptor explains the mechanisms for gain- and loss-of function mutations
Nat Commun, 13, 2022
|
|
7VMR
 
 | | Structure of recombinant RyR2 mutant K4593A (EGTA dataset) | | Descriptor: | Peptidyl-prolyl cis-trans isomerase FKBP1B, Ryanodine receptor 2, ZINC ION | | Authors: | Kobayashi, T, Tsutsumi, A, Kurebayashi, N, Kodama, M, Kikkawa, M, Murayama, T, Ogawa, H. | | Deposit date: | 2021-10-09 | | Release date: | 2022-08-10 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Molecular basis for gating of cardiac ryanodine receptor explains the mechanisms for gain- and loss-of function mutations.
Nat Commun, 13, 2022
|
|
7U0T
 
 | | Crystal Structure of a human Calcineurin A - Calcineurin B fusion bound to FKBP12 and FK-520 | | Descriptor: | (3S,4R,5S,8R,9E,12S,14S,15R,16S,18R,19R,22R,26aS)-8-ethyl-5,19-dihydroxy-3-{(1E)-1-[(1R,3R,4R)-4-hydroxy-3-methoxycyclohexyl]prop-1-en-2-yl}-14,16-dimethoxy-4,10,12,18-tetramethyl-5,6,8,11,12,13,14,15,16,17,18,19,24,25,26,26a-hexadecahydro-3H-15,19-epoxypyrido[2,1-c][1,4]oxazacyclotricosine-1,7,20,21(4H,23H)-tetrone, 1,2-ETHANEDIOL, CALCIUM ION, ... | | Authors: | Fox III, D, Mayclin, S.J, DeBouver, N.D, Hoy, M.J, Heitman, J, Lorimer, D.D, Horanyi, P.S, Edwards, T.E, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2022-02-18 | | Release date: | 2022-08-03 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structure-Guided Synthesis of FK506 and FK520 Analogs with Increased Selectivity Exhibit In Vivo Therapeutic Efficacy against Cryptococcus.
Mbio, 13, 2022
|
|
7U9Q
 
 | | Structure of PKA phosphorylated human RyR2 in the closed state | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Peptidyl-prolyl cis-trans isomerase FKBP1B, Ryanodine receptor 2, ... | | Authors: | Miotto, M.C, Marks, A.R. | | Deposit date: | 2022-03-11 | | Release date: | 2022-08-03 | | Method: | ELECTRON MICROSCOPY (3.11 Å) | | Cite: | Structural analyses of human ryanodine receptor type 2 channels reveal the mechanisms for sudden cardiac death and treatment.
Sci Adv, 8, 2022
|
|
7U9X
 
 | |
7U9T
 
 | |
