3UEI
 
 | | Crystal structure of human Survivin E65A mutant | | Descriptor: | Baculoviral IAP repeat-containing protein 5, ZINC ION | | Authors: | Niedzialkowska, E, Porebski, P.J, Wang, F, Higgins, J.M, Stukenberg, P.T, Minor, W. | | Deposit date: | 2011-10-30 | | Release date: | 2012-03-07 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Molecular basis for phosphospecific recognition of histone H3 tails by Survivin paralogues at inner centromeres.
Mol Biol Cell, 23, 2012
|
|
3UEH
 
 | | Crystal structure of human Survivin H80A mutant | | Descriptor: | 1,2-ETHANEDIOL, Baculoviral IAP repeat-containing protein 5, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Niedzialkowska, E, Porebski, P.J, Wang, F, Higgins, J.M, Stukenberg, P.T, Minor, W. | | Deposit date: | 2011-10-30 | | Release date: | 2012-03-07 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Molecular basis for phosphospecific recognition of histone H3 tails by Survivin paralogues at inner centromeres.
Mol Biol Cell, 23, 2012
|
|
3UIG
 
 | |
5BT1
 
 | | histone chaperone Hif1 playing with histone H2A-H2B dimer | | Descriptor: | HAT1-interacting factor 1, Histone H2A.1, Histone H2B.1 | | Authors: | Liu, H, Zhang, M, Gao, Y, Teng, M, Niu, L. | | Deposit date: | 2015-06-02 | | Release date: | 2016-10-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | Structural Insights into the Association of Hif1 with Histones H2A-H2B Dimer and H3-H4 Tetramer
Structure, 24, 2016
|
|
2F5K
 
 | |
3O35
 
 | |
7VRF
 
 | | Crystal structure of Oxpecker chromodomain in complex with H3K9me3 | | Descriptor: | H3K9me3, Oxpecker | | Authors: | Huang, Y, Jin, Z, Yu, B. | | Deposit date: | 2021-10-22 | | Release date: | 2022-10-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural insights into the chromodomain of Oxpecker in complex with histone H3 lysine 9 trimethylation reveal a transposon silencing mechanism by heterodimerization.
Biochem.Biophys.Res.Commun., 652, 2023
|
|
5Y2L
 
 | |
2R0Y
 
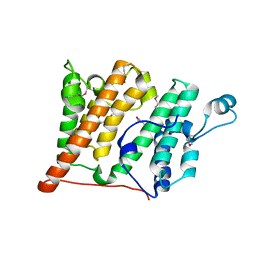 | | Structure of the Rsc4 tandem bromodomain in complex with an acetylated H3 peptide | | Descriptor: | Chromatin structure-remodeling complex protein RSC4, Histone H3 peptide | | Authors: | VanDemark, A.P, Kasten, M.M, Ferris, E, Heroux, A, Hill, C.P, Cairns, B.R. | | Deposit date: | 2007-08-21 | | Release date: | 2007-10-30 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Autoregulation of the rsc4 tandem bromodomain by gcn5 acetylation.
Mol.Cell, 27, 2007
|
|
8JWS
 
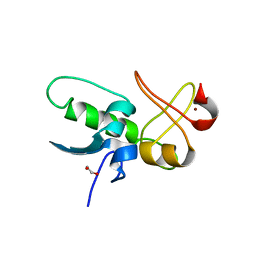 | | ePHD domain of PHD Finger Protein 7 (PHF7) | | Descriptor: | 1,2-ETHANEDIOL, PHD finger protein 7, ZINC ION | | Authors: | Bang, I, Lee, H.S, Choi, H.-J. | | Deposit date: | 2023-06-29 | | Release date: | 2023-12-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Molecular basis for PHF7-mediated ubiquitination of histone H3.
Genes Dev., 37, 2023
|
|
8JWJ
 
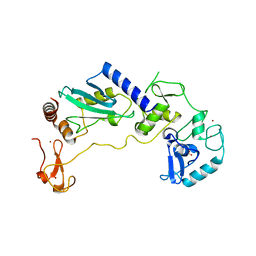 | | PHD Finger Protein 7 (PHF7) in complex with UBE2D2 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, GLYCEROL, PHD finger protein 7, ... | | Authors: | Lee, H.S, Bang, I, Choi, H.-J. | | Deposit date: | 2023-06-29 | | Release date: | 2023-12-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.96 Å) | | Cite: | Molecular basis for PHF7-mediated ubiquitination of histone H3.
Genes Dev., 37, 2023
|
|
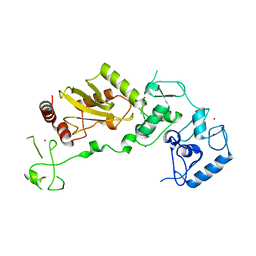
8JWU
 
 | |
3SOU
 
 | |
1QSR
 
 | | CRYSTAL STRUCTURE OF TETRAHYMENA GCN5 WITH BOUND ACETYL-COENZYME A | | Descriptor: | ACETYL COENZYME *A, TGCN5 HISTONE ACETYL TRANSFERASE | | Authors: | Rojas, J.R, Trievel, R.C, Zhou, J, Mo, Y, Li, X, Berger, S.L, David Allis, C, Marmorstein, R. | | Deposit date: | 1999-06-23 | | Release date: | 1999-09-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of Tetrahymena GCN5 bound to coenzyme A and a histone H3 peptide.
Nature, 401, 1999
|
|
1QST
 
 | | CRYSTAL STRUCTURE OF TETRAHYMENA GCN5 | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, TGCN5 HISTONE ACETYL TRANSFERASE | | Authors: | Rojas, J.R, Trievel, R.C, Zhou, J, Mo, Y, Li, X, Berger, S.L, David Allis, C, Marmorstein, R. | | Deposit date: | 1999-06-23 | | Release date: | 1999-09-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of Tetrahymena GCN5 bound to coenzyme A and a histone H3 peptide.
Nature, 401, 1999
|
|
6NHQ
 
 | |
6NHR
 
 | |
6NHP
 
 | |
6OI0
 
 | | Crystal structure of human WDR5 in complex with L-arginine | | Descriptor: | ARGININE, GLYCEROL, SULFATE ION, ... | | Authors: | Lorton, B.M, Harijan, R.K, Burgos, E, Bonanno, J.B, Almo, S.C, Shechter, D. | | Deposit date: | 2019-04-08 | | Release date: | 2020-04-01 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | A Binary Arginine Methylation Switch on Histone H3 Arginine 2 Regulates Its Interaction with WDR5.
Biochemistry, 59, 2020
|
|
6OFZ
 
 | | Crystal structure of human WDR5 | | Descriptor: | WD repeat-containing protein 5 | | Authors: | Lorton, B.M, Harijan, R.K, Burgos, E, Bonanno, J.B, Almo, S.C, Shechter, D. | | Deposit date: | 2019-04-01 | | Release date: | 2020-04-01 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | A Binary Arginine Methylation Switch on Histone H3 Arginine 2 Regulates Its Interaction with WDR5.
Biochemistry, 59, 2020
|
|
6OI1
 
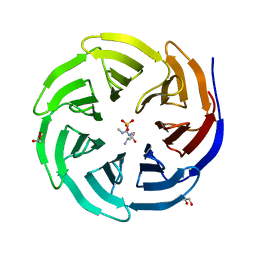 | | Crystal structure of human WDR5 in complex with monomethyl L-arginine | | Descriptor: | (2S)-2-amino-5-[(N-methylcarbamimidoyl)amino]pentanoic acid, GLYCEROL, SULFATE ION, ... | | Authors: | Lorton, B.M, Harijan, R.K, Burgos, E, Bonanno, J.B, Almo, S.C, Shechter, D. | | Deposit date: | 2019-04-08 | | Release date: | 2020-04-01 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | A Binary Arginine Methylation Switch on Histone H3 Arginine 2 Regulates Its Interaction with WDR5.
Biochemistry, 59, 2020
|
|
2I7K
 
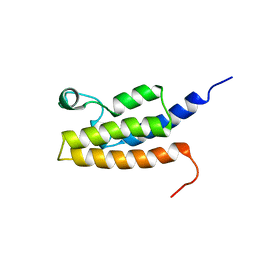 | | Solution Structure of the Bromodomain of Human BRD7 Protein | | Descriptor: | Bromodomain-containing protein 7 | | Authors: | Sun, H, Liu, J, Zhang, J, Huang, H, Wu, J, Shi, Y. | | Deposit date: | 2006-08-31 | | Release date: | 2007-07-10 | | Last modified: | 2022-03-09 | | Method: | SOLUTION NMR | | Cite: | Solution structure of BRD7 bromodomain and its interaction with acetylated peptides from histone H3 and H4
Biochem.Biophys.Res.Commun., 358, 2007
|
|
7ZMX
 
 | |
5SZB
 
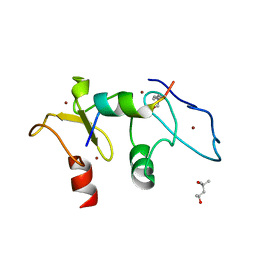 | | Structure of human Dpf3 double-PHD domain bound to histone H3 tail peptide with acetylated K14 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Histone H3 tail peptide, ZINC ION, ... | | Authors: | Singh, N, Local, A, Shiau, A, Ren, B, Corbett, K.D. | | Deposit date: | 2016-08-13 | | Release date: | 2017-08-16 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Identification of H3K4me1-associated proteins at mammalian enhancers.
Nat. Genet., 50, 2018
|
|
5U7G
 
 | | Crystal Structure of the Catalytic Core of CBP | | Descriptor: | CREB-binding protein, ZINC ION | | Authors: | Park, S, Stanfield, R.L, Martinez-Yamout, M.M, Dyson, H.J, Wilson, I.A, Wright, P.E. | | Deposit date: | 2016-12-12 | | Release date: | 2017-06-21 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.401 Å) | | Cite: | Role of the CBP catalytic core in intramolecular SUMOylation and control of histone H3 acetylation.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
