4EQ3
 
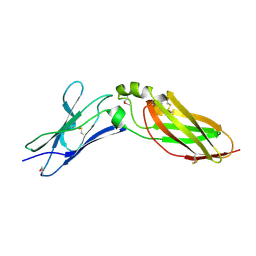 | | Crystal Structure Analysis of Selenomethionine (Se-Met) Substituted Chicken Interferon Gamma Receptor Alpha Chain | | Descriptor: | Interferon gamma receptor 1 | | Authors: | Ping, Z, Qi, J, Lu, G, Shi, Y, Wang, X, Gao, G.F, Wang, M. | | Deposit date: | 2012-04-18 | | Release date: | 2013-04-24 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.001 Å) | | Cite: | Crystal structure of the interferon gamma receptor alpha chain from chicken reveals an undetected extra helix compared with the human counterparts.
J.Interferon Cytokine Res., 34, 2014
|
|
4EQ2
 
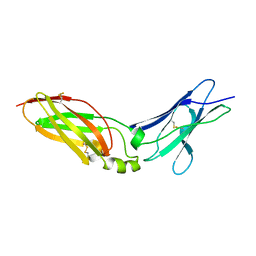 | | Crystal Structure Analysis of Chicken Interferon Gamma Receptor Alpha Chain | | Descriptor: | Interferon gamma receptor 1 | | Authors: | Ping, Z, Qi, J, Lu, G, Shi, Y, Wang, X, Gao, G.F, Wang, M. | | Deposit date: | 2012-04-18 | | Release date: | 2013-04-24 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.502 Å) | | Cite: | Crystal structure of the interferon gamma receptor alpha chain from chicken reveals an undetected extra helix compared with the human counterparts.
J.Interferon Cytokine Res., 34, 2014
|
|
2LAG
 
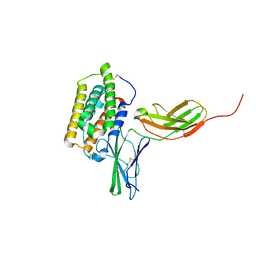 | | Structure of the 44 kDa complex of interferon-alpha2 with the extracellular part of IFNAR2 obtained by 2D-double difference NOESY | | Descriptor: | Interferon alpha-2, Interferon alpha/beta receptor 2 | | Authors: | Nudelman, I, Akabayov, S.R, Scherf, T, Anglister, J. | | Deposit date: | 2011-03-13 | | Release date: | 2011-08-17 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Observation of Intermolecular Interactions in Large Protein Complexes by 2D-Double Difference Nuclear Overhauser Enhancement Spectroscopy: Application to the 44 kDa Interferon-Receptor Complex.
J.Am.Chem.Soc., 133, 2011
|
|
5UG6
 
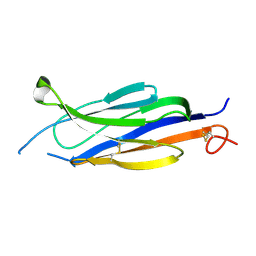 | | Perforin C2 Domain - T431D | | Descriptor: | IODIDE ION, Perforin-1 | | Authors: | Law, R.H.P, Conroy, P.J, Voskoboinik, I, Whisstock, J.C. | | Deposit date: | 2017-01-07 | | Release date: | 2018-02-07 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Perforin proteostasis is regulated through its C2 domain: supra-physiological cell death mediated by T431D-perforin.
Cell Death Differ., 25, 2018
|
|
6F1E
 
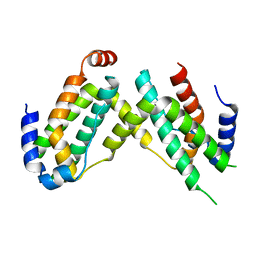 | | Crystal structure of olive flounder [Paralichthys olivaceus] interferon gamma at 2.3 Angstrom resolution | | Descriptor: | Interferon gamma | | Authors: | Kolenko, P, Kolarova, L, Zahradnik, J, Schneider, B. | | Deposit date: | 2017-11-21 | | Release date: | 2018-05-23 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.296 Å) | | Cite: | Interferons type II and their receptors R1 and R2 in fish species: Evolution, structure, and function.
Fish Shellfish Immunol., 79, 2018
|
|
4CRN
 
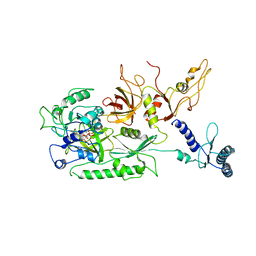 | | Cryo-EM of a pretermination complex with eRF1 and eRF3 | | Descriptor: | ERF1 IN RIBOSOME-BOUND ERF1-ERF3-GDPNP COMPLEX, ERF3 IN RIBOSOME BOUND ERF1-ERF3-GDPNP COMPLEX, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER | | Authors: | Preis, A, Heuer, A, Barrio-Garcia, C, Hauser, A, Eyler, D, Berninghausen, O, Green, R, Becker, T, Beckmann, R. | | Deposit date: | 2014-02-28 | | Release date: | 2014-07-23 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (9.1 Å) | | Cite: | Cryoelectron Microscopic Structures of Eukaryotic Translation Termination Complexes Containing Erf1-Erf3 or Erf1-Abce1.
Cell Rep., 8, 2014
|
|
5UG7
 
 | | Calcium bound Perforin C2 Domain - T431D | | Descriptor: | CALCIUM ION, Perforin-1 | | Authors: | Law, R.H.P, Conroy, P.J, Voskoboinik, I, Whisstock, J.C. | | Deposit date: | 2017-01-07 | | Release date: | 2018-02-07 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Perforin proteostasis is regulated through its C2 domain: supra-physiological cell death mediated by T431D-perforin.
Cell Death Differ., 25, 2018
|
|
7JSL
 
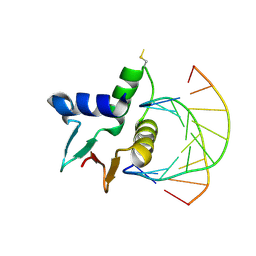 | |
7JSA
 
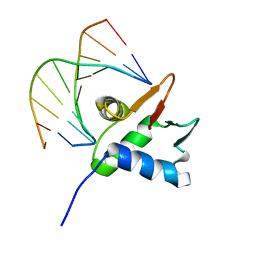 | |
7PAG
 
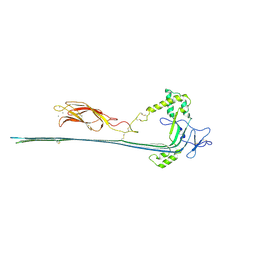 | | The pore conformation of lymphocyte perforin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Perforin-1 | | Authors: | Ivanova, M.E, Lukoyanova, N, Malhotra, S, Topf, M, Trapani, J.A, Voskoboinik, I, Saibil, H.R. | | Deposit date: | 2021-07-29 | | Release date: | 2022-02-16 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | The pore conformation of lymphocyte perforin.
Sci Adv, 8, 2022
|
|
3J5Y
 
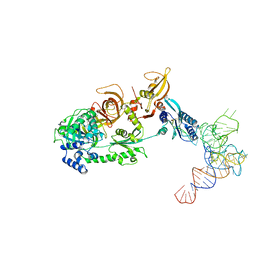 | | Structure of the mammalian ribosomal pre-termination complex associated with eRF1-eRF3-GDPNP | | Descriptor: | 5'-R(*AP*UP*UP*GP*UP*AP*AP*AP*AP*A)-3', Eukaryotic peptide chain release factor GTP-binding subunit ERF3A, Eukaryotic peptide chain release factor subunit 1, ... | | Authors: | des Georges, A, Hashem, Y, Unbehaun, A, Grassucci, R.A, Taylor, D, Hellen, C.U.T, Pestova, T.V, Frank, J. | | Deposit date: | 2013-11-21 | | Release date: | 2013-12-25 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (9.7 Å) | | Cite: | Structure of the mammalian ribosomal pre-termination complex associated with eRF1*eRF3*GDPNP.
Nucleic Acids Res., 42, 2014
|
|
3E1Y
 
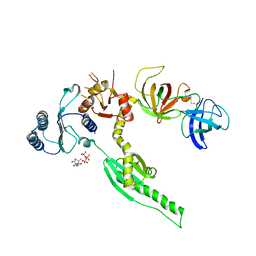 | | Crystal structure of human eRF1/eRF3 complex | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Eukaryotic peptide chain release factor GTP-binding subunit ERF3A, Eukaryotic peptide chain release factor subunit 1 | | Authors: | Cheng, Z, Lim, M, Kong, C, Song, H. | | Deposit date: | 2008-08-05 | | Release date: | 2009-05-19 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Structural insights into eRF3 and stop codon recognition by eRF1
Genes Dev., 23, 2009
|
|
2MOZ
 
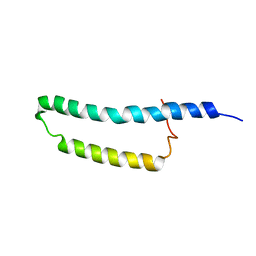 | | Structure of the Membrane Protein MerF, a Bacterial Mercury Transporter, Improved by the Inclusion of Chemical Shift Anisotropy Constraints | | Descriptor: | MerF | | Authors: | Tian, Y, Lu, G.J, Marassi, F.M, Opella, S.J, Membrane Protein Structures by Solution NMR (MPSbyNMR) | | Deposit date: | 2014-05-07 | | Release date: | 2014-07-30 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure of the membrane protein MerF, a bacterial mercury transporter, improved by the inclusion of chemical shift anisotropy constraints.
J.Biomol.Nmr, 60, 2014
|
|
1ITF
 
 | | INTERFERON ALPHA-2A, NMR, 24 STRUCTURES | | Descriptor: | INTERFERON ALPHA-2A | | Authors: | Klaus, W, Gsell, B, Labhardt, A.M, Wipf, B, Senn, H. | | Deposit date: | 1997-08-22 | | Release date: | 1997-12-03 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | The three-dimensional high resolution structure of human interferon alpha-2a determined by heteronuclear NMR spectroscopy in solution.
J.Mol.Biol., 274, 1997
|
|
1OIW
 
 | | X-ray structure of the small G protein Rab11a in complex with GTPgammaS | | Descriptor: | 5'-GUANOSINE-DIPHOSPHATE-MONOTHIOPHOSPHATE, MAGNESIUM ION, RAS-RELATED PROTEIN RAB-11A | | Authors: | Pasqualato, S, Senic-Matuglia, F, Renault, L, Goud, B, Salamero, J, Cherfils, J. | | Deposit date: | 2003-06-26 | | Release date: | 2004-01-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | The Structural Gdp/GTP Cycle of Rab11 Reveals a Novel Interface Involved in the Dynamics of Recycling Endosomes
J.Biol.Chem., 279, 2004
|
|
1OIV
 
 | | X-ray structure of the small G protein Rab11a in complex with GDP | | Descriptor: | 1,2-ETHANEDIOL, GUANOSINE-5'-DIPHOSPHATE, RAS-RELATED PROTEIN RAB-11A, ... | | Authors: | Pasqualato, S, Senic-Matuglia, F, Renault, L, Goud, B, Salamero, J, Cherfils, J. | | Deposit date: | 2003-06-26 | | Release date: | 2004-01-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | The Structural Gdp/GTP Cycle of Rab11 Reveals a Novel Interface Involved in the Dynamics of Recycling Endosomes
J.Biol.Chem., 279, 2004
|
|
4L9Z
 
 | | Crystal Structure of Rhodobacter sphaeroides malyl-CoA lyase in complex with magnesium, oxalate, and CoA | | Descriptor: | COENZYME A, MAGNESIUM ION, Malyl-CoA lyase, ... | | Authors: | Zarzycki, J, Kerfeld, C.A. | | Deposit date: | 2013-06-18 | | Release date: | 2013-12-04 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.011 Å) | | Cite: | The crystal structures of the tri-functional Chloroflexus aurantiacus and bi-functional Rhodobacter sphaeroides malyl-CoA lyases and comparison with CitE-like superfamily enzymes and malate synthases.
Bmc Struct.Biol., 13, 2013
|
|
4L80
 
 | | Crystal Structure of Chloroflexus aurantiacus malyl-CoA lyase in complex with magnesium, oxalate, and propionyl-CoA | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, HpcH/HpaI aldolase, MAGNESIUM ION, ... | | Authors: | Zarzycki, J, Kerfeld, C.A. | | Deposit date: | 2013-06-15 | | Release date: | 2013-12-04 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.008 Å) | | Cite: | The crystal structures of the tri-functional Chloroflexus aurantiacus and bi-functional Rhodobacter sphaeroides malyl-CoA lyases and comparison with CitE-like superfamily enzymes and malate synthases.
Bmc Struct.Biol., 13, 2013
|
|
4L9Y
 
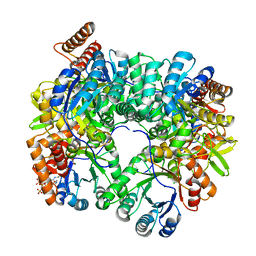 | | Crystal Structure of Rhodobacter sphaeroides malyl-CoA lyase in complex with magnesium, glyoxylate, and propionyl-CoA | | Descriptor: | CHLORIDE ION, GLYOXYLIC ACID, MAGNESIUM ION, ... | | Authors: | Zarzycki, J, Kerfeld, C.A. | | Deposit date: | 2013-06-18 | | Release date: | 2013-12-04 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.102 Å) | | Cite: | The crystal structures of the tri-functional Chloroflexus aurantiacus and bi-functional Rhodobacter sphaeroides malyl-CoA lyases and comparison with CitE-like superfamily enzymes and malate synthases.
Bmc Struct.Biol., 13, 2013
|
|
4L7Z
 
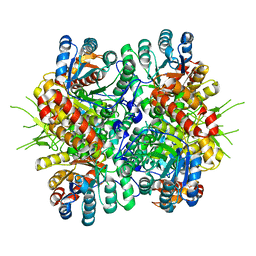 | | Crystal Structure of Chloroflexus aurantiacus malyl-CoA lyase | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, HpcH/HpaI aldolase | | Authors: | Zarzycki, J, Kerfeld, C.A. | | Deposit date: | 2013-06-14 | | Release date: | 2013-12-04 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.502 Å) | | Cite: | The crystal structures of the tri-functional Chloroflexus aurantiacus and bi-functional Rhodobacter sphaeroides malyl-CoA lyases and comparison with CitE-like superfamily enzymes and malate synthases.
Bmc Struct.Biol., 13, 2013
|
|
3DSH
 
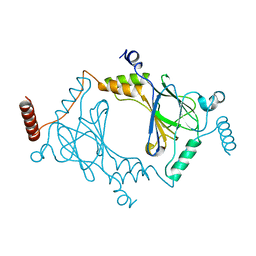 | | Crystal structure of dimeric interferon regulatory factor 5 (IRF-5) transactivation domain | | Descriptor: | Interferon regulatory factor 5 | | Authors: | Chen, W, Lam, S.S, Srinath, H, Jiang, Z, Correia, J.J, Schiffer, C, Fitzgerald, K.A, Lin, K, Royer Jr, W.E. | | Deposit date: | 2008-07-12 | | Release date: | 2008-10-07 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Insights into interferon regulatory factor activation from the crystal structure of dimeric IRF5.
Nat.Struct.Mol.Biol., 15, 2008
|
|
2VLG
 
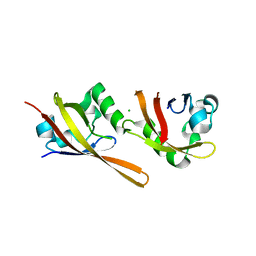 | | KinA PAS-A domain, homodimer | | Descriptor: | ACETATE ION, CHLORIDE ION, SPORULATION KINASE A | | Authors: | Lee, J, Tomchick, D.R, Brautigam, C.A, Machius, M, Kort, R, Hellingwerf, K.J, Gardner, K.H. | | Deposit date: | 2008-01-14 | | Release date: | 2008-03-18 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Changes at the Kina Pas-A Dimerization Interface Influence Histidine Kinase Function.
Biochemistry, 47, 2008
|
|
4Z5R
 
 | | Rontalizumab Fab bound to Interferon-a2 | | Descriptor: | Interferon alpha-2, SULFATE ION, anti-IFN-a antibody rontalizumab heavy chain modules VH and CH1 (Fab), ... | | Authors: | Eigenbrot, C, Maurer, B, Bosanac, I. | | Deposit date: | 2015-04-02 | | Release date: | 2015-07-08 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural basis of the broadly neutralizing anti-interferon-alpha antibody rontalizumab.
Protein Sci., 24, 2015
|
|
4Y1S
 
 | | Structural basis for Ca2+-mediated interaction of the perforin C2 domain with lipid membranes | | Descriptor: | CALCIUM ION, Perforin-1 | | Authors: | Conroy, P.J, Yagi, H, Whisstock, J.C, Norton, R.S. | | Deposit date: | 2015-02-09 | | Release date: | 2015-09-02 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.611 Å) | | Cite: | Structural Basis for Ca2+-mediated Interaction of the Perforin C2 Domain with Lipid Membranes.
J.Biol.Chem., 290, 2015
|
|
4Y1T
 
 | | Structural basis for Ca2+-mediated interaction of the perforin C2 domain with lipid membranes | | Descriptor: | CALCIUM ION, Perforin-1 | | Authors: | Conroy, P.J, Yagi, H, Whisstock, J.C, Norton, R.S. | | Deposit date: | 2015-02-09 | | Release date: | 2015-09-02 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.666 Å) | | Cite: | Structural Basis for Ca2+-mediated Interaction of the Perforin C2 Domain with Lipid Membranes.
J.Biol.Chem., 290, 2015
|
|
