3E9P
 
 | |
6NIT
 
 | |
6MDZ
 
 | |
6MFR
 
 | |
6MFN
 
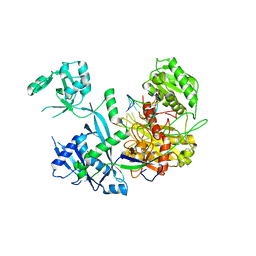 | |
8VAJ
 
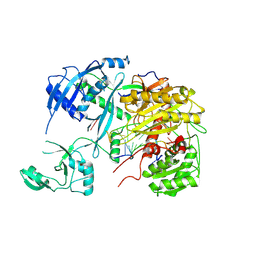 | | Human Argonaute3 bound to cityRNA and target RNA | | Descriptor: | Protein argonaute-3, RNA (5'-R(P*(SRA)P*AP*GP*CP*AP*CP*UP*UP*UP*AP*AP*A)-3'), RNA (5'-R(P*UP*AP*AP*AP*GP*UP*GP*CP*UP*UP*AP*G)-3') | | Authors: | Zhang, H, Sim, G, Adhav, V.A, Nakanishi, K. | | Deposit date: | 2023-12-11 | | Release date: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.45 Å) | | Cite: | Target cleavage and gene silencing by Argonautes with cityRNAs.
Cell Rep, 43, 2024
|
|
2CZJ
 
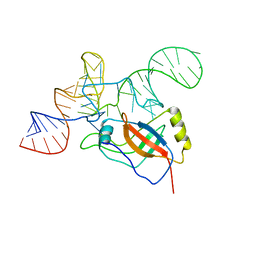 | | Crystal structure of the tRNA domain of tmRNA from Thermus thermophilus HB8 | | Descriptor: | SsrA-binding protein, tmRNA (63-MER) | | Authors: | Bessho, Y, Shibata, R, Sekine, S, Murayama, K, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-07-13 | | Release date: | 2006-10-31 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.01 Å) | | Cite: | Structural basis for functional mimicry of long-variable-arm tRNA by transfer-messenger RNA.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
2NUF
 
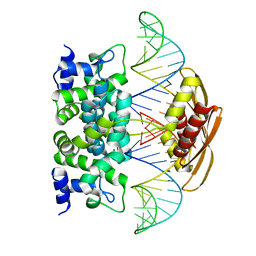 | | Crystal structure of RNase III from Aquifex aeolicus complexed with ds-RNA at 2.5-Angstrom Resolution | | Descriptor: | 28-MER, MAGNESIUM ION, Ribonuclease III | | Authors: | Gan, J.H, Shaw, G, Tropea, J.E, Waugh, D.S, Court, D.L, Ji, X. | | Deposit date: | 2006-11-09 | | Release date: | 2007-11-20 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A stepwise model for double-stranded RNA processing by ribonuclease III.
Mol.Microbiol., 67, 2007
|
|
8VU0
 
 | |
2NUG
 
 | | Crystal structure of RNase III from Aquifex aeolicus complexed with ds-RNA at 1.7-Angstrom Resolution | | Descriptor: | 5'-R(P*AP*AP*GP*GP*UP*CP*AP*UP*UP*CP*G)-3', 5'-R(P*AP*GP*UP*GP*GP*CP*CP*UP*UP*GP*C)-3', MAGNESIUM ION, ... | | Authors: | Gan, J.H, Shaw, G, Tropea, J.E, Waugh, D.S, Court, D.L, Ji, X. | | Deposit date: | 2006-11-09 | | Release date: | 2007-11-20 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A stepwise model for double-stranded RNA processing by ribonuclease III.
Mol.Microbiol., 67, 2007
|
|
2NUE
 
 | | Crystal structure of RNase III from Aquifex aeolicus complexed with ds-RNA at 2.9-Angstrom Resolution | | Descriptor: | 46-MER, Ribonuclease III | | Authors: | Gan, J.H, Shaw, G, Tropea, J.E, Waugh, D.S, Court, D.L, Ji, X. | | Deposit date: | 2006-11-09 | | Release date: | 2007-11-20 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | A stepwise model for double-stranded RNA processing by ribonuclease III.
Mol.Microbiol., 67, 2007
|
|
8CDP
 
 | | Cryo-EM structure of the RESC1-RESC2 complex | | Descriptor: | Guide_RNA_associated_protein_-_putative, Mitochondrial guide RNA binding complex subunit 2 | | Authors: | Dolce, L.G, Weis, F, Kowalinski, E. | | Deposit date: | 2023-01-31 | | Release date: | 2023-03-29 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural basis for guide RNA selection by the RESC1-RESC2 complex.
Nucleic Acids Res., 51, 2023
|
|
4U37
 
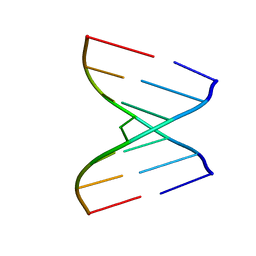 | | Native 7mer-RNA duplex | | Descriptor: | RNA (5'-R(*GP*GP*AP*GP*CP*UP*A)-3'), RNA (5'-R(*UP*AP*GP*CP*UP*CP*C)-3') | | Authors: | Sheng, J, Larsen, A, Heuberger, B, Blain, J.C, Szostak, J.W. | | Deposit date: | 2014-07-18 | | Release date: | 2014-08-13 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal Structure Studies of RNA Duplexes Containing s(2)U:A and s(2)U:U Base Pairs.
J.Am.Chem.Soc., 136, 2014
|
|
4A3I
 
 | | RNA Polymerase II binary complex with DNA | | Descriptor: | 5'-D(*GP*GP*CP*AP*CP*AP*AP*CP*TP*GP*CP*GP*GP*CP*T)-3', DNA-DIRECTED RNA POLYMERASE II SUBUNIT RPB1, DNA-DIRECTED RNA POLYMERASE II SUBUNIT RPB11, ... | | Authors: | Cheung, A.C.M, Sainsbury, S, Cramer, P. | | Deposit date: | 2011-09-30 | | Release date: | 2012-02-08 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Structural basis of initial RNA polymerase II transcription.
EMBO J., 30, 2011
|
|
4V34
 
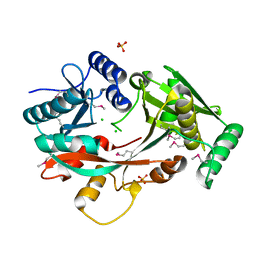 | | The Structure of A-PGS from Pseudomonas aeruginosa (SeMet derivative) | | Descriptor: | ALANYL-TRNA-DEPENDENT L-ALANYL- PHOPHATIDYLGLYCEROL SYNTHASE, CHLORIDE ION, SULFATE ION | | Authors: | Krausze, J, Hebecker, S, Hasenkampf, T, Heinz, D.W, Moser, J. | | Deposit date: | 2014-10-16 | | Release date: | 2015-08-19 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structures of Two Bacterial Resistance Factors Mediating tRNA-Dependent Aminoacylation of Phosphatidylglycerol with Lysine or Alanine.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
5M59
 
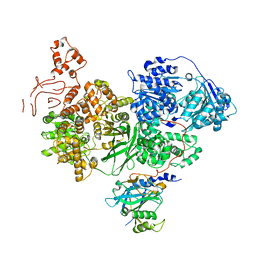 | | Crystal structure of Chaetomium thermophilum Brr2 helicase core in complex with Prp8 Jab1 domain | | Descriptor: | ACETATE ION, Pre-mRNA splicing helicase-like protein, Putative pre-mRNA splicing factor | | Authors: | Absmeier, E, Becke, C, Wollenhaupt, J, Santos, K.F, Wahl, M.C. | | Deposit date: | 2016-10-20 | | Release date: | 2016-12-21 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Interplay of cis- and trans-regulatory mechanisms in the spliceosomal RNA helicase Brr2.
Cell Cycle, 16, 2017
|
|
8TG5
 
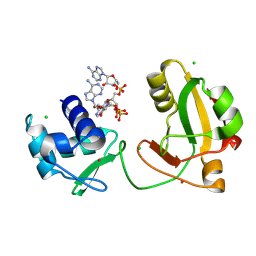 | | tRNA 2'-phosphotransferase (Tpt1) from Pyrococcus horikoshii in complex with branched 2'-PO4 RNA | | Descriptor: | (2S,3R,4R,5S)-2-(6-amino-9H-purin-9-yl)-4-{[(S)-{[(2R,3S,4R,5S)-5-(6-amino-9H-purin-9-yl)-3,4-dihydroxyoxolan-2-yl]methoxy}(hydroxy)phosphoryl]oxy}-5-({[(S)-{[(2R,3R,4S,5R)-5-(6-amino-9H-purin-9-yl)-4-hydroxy-2-(hydroxymethyl)oxolan-3-yl]oxy}(hydroxy)phosphoryl]oxy}methyl)oxolan-3-yl dihydrogen phosphate (non-preferred name), CHLORIDE ION, POTASSIUM ION, ... | | Authors: | Jacewicz, A, Dantuluri, S, Shuman, S. | | Deposit date: | 2023-07-12 | | Release date: | 2024-06-05 | | Last modified: | 2024-06-26 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for Tpt1-catalyzed 2'-PO 4 transfer from RNA and NADP(H) to NAD.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
1I6H
 
 | | RNA POLYMERASE II ELONGATION COMPLEX | | Descriptor: | 5'-D(P*AP*AP*AP*TP*GP*CP*CP*TP*GP*GP*TP*CP*T)-3', 5'-R(P*GP*AP*CP*CP*AP*GP*GP*CP*A)-3', DNA-DIRECTED RNA POLYMERASE II 13.6KD POLYPEPTIDE, ... | | Authors: | Gnatt, A.L, Cramer, P, Kornberg, R.D. | | Deposit date: | 2001-03-02 | | Release date: | 2001-04-23 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structural basis of transcription: an RNA polymerase II elongation complex at 3.3 A resolution.
Science, 292, 2001
|
|
7W0A
 
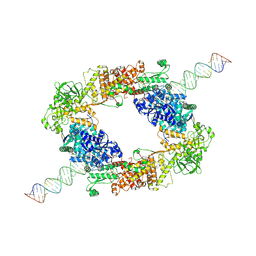 | | dmDicer2-LoqsPD-dsRNA Dimer status | | Descriptor: | Dicer-2, isoform A, Loquacious, ... | | Authors: | Su, S, Wang, J, Wang, H.W, Ma, J. | | Deposit date: | 2021-11-18 | | Release date: | 2022-04-27 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (3.12 Å) | | Cite: | Structural insights into dsRNA processing by Drosophila Dicer-2-Loqs-PD.
Nature, 607, 2022
|
|
8TG4
 
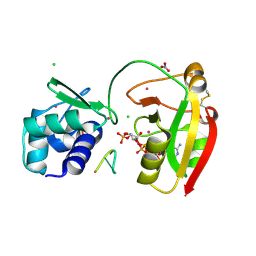 | | tRNA 2'-phosphotransferase (Tpt1) from Aeropyrum pernix in complex with ADP-ribose-2"-phosphate and 2'-OH RNA | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, NITRATE ION, ... | | Authors: | Jacewicz, A, Dantuluri, S, Shuman, S. | | Deposit date: | 2023-07-12 | | Release date: | 2023-11-08 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | Structural basis for Tpt1-catalyzed 2'-PO 4 transfer from RNA and NADP(H) to NAD.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
7W0C
 
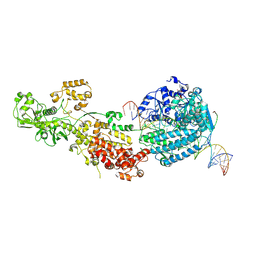 | | Dicer2-Loqs-PD-dsRNA complex at early-translocation state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Dicer-2, isoform A, ... | | Authors: | Su, S, Wang, J, Wang, H.W, Ma, J. | | Deposit date: | 2021-11-18 | | Release date: | 2022-04-27 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (3.93 Å) | | Cite: | Structural insights into dsRNA processing by Drosophila Dicer-2-Loqs-PD.
Nature, 607, 2022
|
|
7ZNK
 
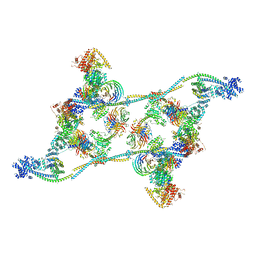 | | Structure of an endogenous human TREX complex bound to mRNA | | Descriptor: | RNA, Spliceosome RNA helicase DDX39B, THO complex subunit 1, ... | | Authors: | Pacheco-Fiallos, F.B, Vorlaender, M.K, Plaschka, C. | | Deposit date: | 2022-04-21 | | Release date: | 2023-05-03 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | mRNA recognition and packaging by the human transcription-export complex.
Nature, 616, 2023
|
|
7F3E
 
 | | Cryo-EM structure of the minimal protein-only RNase P from Aquifex aeolicus | | Descriptor: | RNA-free ribonuclease P | | Authors: | Teramoto, T, Koyasu, T, Adachi, N, Kawasaki, M, Moriya, T, Numata, T, Senda, T, Kakuta, Y. | | Deposit date: | 2021-06-16 | | Release date: | 2021-08-11 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (3.62 Å) | | Cite: | Minimal protein-only RNase P structure reveals insights into tRNA precursor recognition and catalysis.
J.Biol.Chem., 297, 2021
|
|
7W0D
 
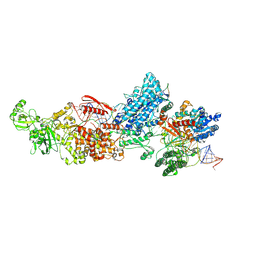 | | Dicer2-LoqsPD-dsRNA complex at mid-translocation state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Dicer-2, isoform A, ... | | Authors: | Su, S, Wang, J, Wang, H.W, Ma, J. | | Deposit date: | 2021-11-18 | | Release date: | 2022-04-27 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (4.18 Å) | | Cite: | Structural insights into dsRNA processing by Drosophila Dicer-2-Loqs-PD.
Nature, 607, 2022
|
|
8IDF
 
 | |
