7R61
 
 | | BTK in complex with 25A | | Descriptor: | 5-{(3S)-1-[(Z)-iminomethyl]piperidin-3-yl}-2-(4-phenoxyphenoxy)pyridine-3-carboxamide, DIMETHYL SULFOXIDE, Tyrosine-protein kinase BTK | | Authors: | Gardberg, A. | | Deposit date: | 2021-06-22 | | Release date: | 2021-10-06 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Discovery of Covalent Bruton's Tyrosine Kinase Inhibitors with Decreased CYP2C8 Inhibitory Activity.
Chemmedchem, 16, 2021
|
|
4TK0
 
 | | Crystal Structure of human Tankyrase 2 in complex with DPQ. | | Descriptor: | 5-[4-(piperidin-1-yl)butoxy]-3,4-dihydroisoquinolin-1(2H)-one, Tankyrase-2, ZINC ION | | Authors: | Qiu, W, Lam, R, Romanov, V, Gordon, R, Gebremeskel, S, Vodsedalek, J, Thompson, C, Beletskaya, I, Battaile, K.P, Pai, E.F, Chirgadze, N.Y. | | Deposit date: | 2014-05-25 | | Release date: | 2014-11-05 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Insights into the binding of PARP inhibitors to the catalytic domain of human tankyrase-2.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
8HZ9
 
 | | Crystal structure of AtHPPD-Y181136 complex | | Descriptor: | 4-hydroxyphenylpyruvate dioxygenase, 5-methyl-6-[(2-methyl-3-oxidanylidene-1H-pyrazol-4-yl)carbonyl]-3-propan-2-yl-1,2,3-benzotriazin-4-one, COBALT (II) ION | | Authors: | Dong, J, Lin, H.-Y, Yang, G.-F. | | Deposit date: | 2023-01-08 | | Release date: | 2023-12-20 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.011 Å) | | Cite: | Crystal structure of AtHPPD-Y181136 complex
To Be Published
|
|
4I89
 
 | | Crystal structure of transthyretin in complex with diflunisal at acidic pH | | Descriptor: | 5-(2,4-DIFLUOROPHENYL)-2-HYDROXY-BENZOIC ACID, Transthyretin | | Authors: | Zanotti, G, Cendron, L, Folli, C, Florio, P, Imbimbo, B.P, Berni, R. | | Deposit date: | 2012-12-03 | | Release date: | 2013-06-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Structural evidence for native state stabilization of a conformationally labile amyloidogenic transthyretin variant by fibrillogenesis inhibitors.
Febs Lett., 587, 2013
|
|
1IE8
 
 | | Crystal Structure Of The Nuclear Receptor For Vitamin D Ligand Binding Domain Bound to KH1060 | | Descriptor: | 5-(2-{1-[1-(4-ETHYL-4-HYDROXY-HEXYLOXY)-ETHYL]-7A-METHYL-OCTAHYDRO-INDEN-4-YLIDENE}-ETHYLIDENE)-4-METHYLENE-CYCLOHEXANE-1,3-DIOL, VITAMIN D3 RECEPTOR | | Authors: | Tocchini-Valentini, G, Rochel, N, Wurtz, J.M, Mitschler, A, Moras, D. | | Deposit date: | 2001-04-09 | | Release date: | 2001-05-16 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Crystal structures of the vitamin D receptor complexed to superagonist 20-epi ligands.
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
4OXY
 
 | | Substrate-binding loop movement with inhibitor PT10 in the tetrameric Mycobacterium tuberculosis enoyl-ACP reductase InhA | | Descriptor: | 5-hexyl-2-(2-nitrophenoxy)phenol, Enoyl-[acyl-carrier-protein] reductase [NADH], NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Li, H.J, Sullivan, T.J, Pan, P, Lai, C.T, Liu, N, Garcia-Diaz, M, Simmerling, C, Tonge, P.J. | | Deposit date: | 2014-02-09 | | Release date: | 2014-04-30 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.3501 Å) | | Cite: | A Structural and Energetic Model for the Slow-Onset Inhibition of the Mycobacterium tuberculosis Enoyl-ACP Reductase InhA.
Acs Chem.Biol., 9, 2014
|
|
3DQR
 
 | | Structure of neuronal NOS D597N/M336V mutant heme domain in complex with a inhibitor (+-)-N1-{cis-4'-[(6"-aminopyridin-2"-yl)methyl]pyrrolidin-3'-yl}ethane-1,2-diamine | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, ACETATE ION, N-{(3S,4S)-4-[(6-aminopyridin-2-yl)methyl]pyrrolidin-3-yl}ethane-1,2-diamine, ... | | Authors: | Igarashi, J, Li, H, Poulos, T.L. | | Deposit date: | 2008-07-09 | | Release date: | 2009-03-31 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structures of constitutive nitric oxide synthases in complex with de novo designed inhibitors.
J.Med.Chem., 52, 2009
|
|
5RVQ
 
 | | PanDDA analysis group deposition -- Crystal structure of SARS-CoV-2 NSP3 macrodomain in complex with ZINC000002508153 | | Descriptor: | 5-methyl-1H-indole-2-carboxylic acid, Non-structural protein 3 | | Authors: | Correy, G.C, Young, I.D, Thompson, M.C, Fraser, J.S. | | Deposit date: | 2020-10-02 | | Release date: | 2020-12-16 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.08 Å) | | Cite: | Fragment binding to the Nsp3 macrodomain of SARS-CoV-2 identified through crystallographic screening and computational docking.
Sci Adv, 7, 2021
|
|
4QEE
 
 | |
4QDW
 
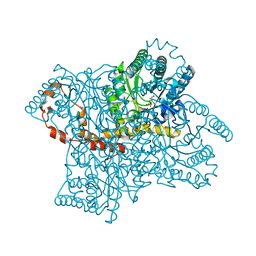 | | Joint X-ray and neutron structure of Streptomyces rubiginosus D-xylose isomerase in complex with two Ni2+ ions and linear L-arabinose | | Descriptor: | L-arabinose, NICKEL (II) ION, Xylose isomerase | | Authors: | Kovalevsky, A.Y, Langan, P. | | Deposit date: | 2014-05-14 | | Release date: | 2014-09-03 | | Last modified: | 2024-02-28 | | Method: | NEUTRON DIFFRACTION (1.8 Å), X-RAY DIFFRACTION | | Cite: | L-Arabinose Binding, Isomerization, and Epimerization by D-Xylose Isomerase: X-Ray/Neutron Crystallographic and Molecular Simulation Study.
Structure, 22, 2014
|
|
5RTQ
 
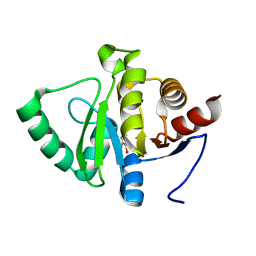 | | PanDDA analysis group deposition -- Crystal structure of SARS-CoV-2 NSP3 macrodomain in complex with ZINC000019015078 | | Descriptor: | 5-bromo-6-methylpyridin-2-amine, Non-structural protein 3 | | Authors: | Correy, G.J, Young, I.D, Thompson, M.C, Fraser, J.S. | | Deposit date: | 2020-09-28 | | Release date: | 2020-12-16 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Fragment binding to the Nsp3 macrodomain of SARS-CoV-2 identified through crystallographic screening and computational docking.
Sci Adv, 7, 2021
|
|
4NKT
 
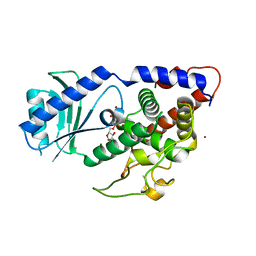 | | Structure of Cid1 in complex with the UTP analog UMPNPP | | Descriptor: | 5'-O-[(S)-hydroxy{[(S)-hydroxy(phosphonooxy)phosphoryl]amino}phosphoryl]uridine, BROMIDE ION, MAGNESIUM ION, ... | | Authors: | Munoz-Tello, P, Gabus, C, Thore, S. | | Deposit date: | 2013-11-13 | | Release date: | 2013-12-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A critical switch in the enzymatic properties of the Cid1 protein deciphered from its product-bound crystal structure.
Nucleic Acids Res., 42, 2014
|
|
4QE5
 
 | |
4Q2V
 
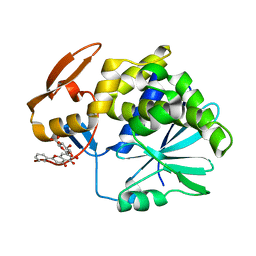 | | Crystal Structure of Ricin A chain complexed with Baicalin inhibitor | | Descriptor: | 5,6-dihydroxy-4-oxo-2-phenyl-4H-chromen-7-yl beta-D-glucopyranosiduronic acid, Ricin | | Authors: | Deng, X, Li, X, Dong, J, Chen, Y. | | Deposit date: | 2014-04-10 | | Release date: | 2015-04-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.198 Å) | | Cite: | Baicalin inhibits the lethality of ricin in mice by inducing protein oligomerization.
J.Biol.Chem., 290, 2015
|
|
4QE4
 
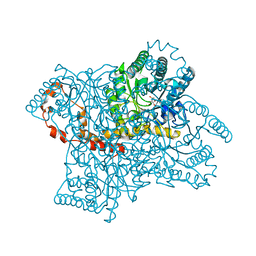 | |
5RVS
 
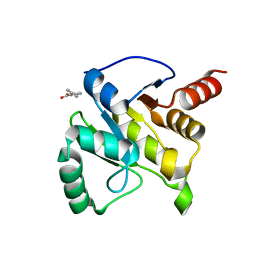 | | PanDDA analysis group deposition -- Crystal structure of SARS-CoV-2 NSP3 macrodomain in complex with ZINC000000159004 | | Descriptor: | 5-phenylpyridine-3-carboxylic acid, Non-structural protein 3 | | Authors: | Correy, G.C, Young, I.D, Thompson, M.C, Fraser, J.S. | | Deposit date: | 2020-10-02 | | Release date: | 2020-12-16 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Fragment binding to the Nsp3 macrodomain of SARS-CoV-2 identified through crystallographic screening and computational docking.
Sci Adv, 7, 2021
|
|
4QEH
 
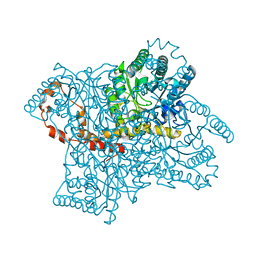 | |
5RTS
 
 | | PanDDA analysis group deposition -- Crystal structure of SARS-CoV-2 NSP3 macrodomain in complex with ZINC000000159004 | | Descriptor: | 5-phenylpyridine-3-carboxylic acid, Non-structural protein 3 | | Authors: | Correy, G.J, Young, I.D, Thompson, M.C, Fraser, J.S. | | Deposit date: | 2020-09-28 | | Release date: | 2020-12-16 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Fragment binding to the Nsp3 macrodomain of SARS-CoV-2 identified through crystallographic screening and computational docking.
Sci Adv, 7, 2021
|
|
4QDP
 
 | | Joint X-ray and neutron structure of Streptomyces rubiginosus D-xylose isomerase in complex with two Cd2+ ions and cyclic beta-L-arabinose | | Descriptor: | CADMIUM ION, Xylose isomerase, beta-L-arabinopyranose | | Authors: | Kovalevsky, A.Y, Langan, P. | | Deposit date: | 2014-05-14 | | Release date: | 2014-09-03 | | Last modified: | 2024-02-28 | | Method: | NEUTRON DIFFRACTION (2 Å), X-RAY DIFFRACTION | | Cite: | L-Arabinose Binding, Isomerization, and Epimerization by D-Xylose Isomerase: X-Ray/Neutron Crystallographic and Molecular Simulation Study.
Structure, 22, 2014
|
|
4QE1
 
 | |
4D7H
 
 | | Structure of Bacillus subtilis nitric oxide synthase in complex with 7-(2-(3-(3-Fluorophenyl(propylamino)ethyl))quinolin-2- amine | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, 7-[2-[3-(3-fluorophenyl)propylamino]ethyl]quinolin-2-amine, CHLORIDE ION, ... | | Authors: | Holden, J.K, Poulos, T.L. | | Deposit date: | 2014-11-25 | | Release date: | 2015-07-01 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Nitric Oxide Synthase as a Target for Methicillin-Resistant Staphylococcus Aureus
Chem.Biol., 22, 2015
|
|
4D7J
 
 | | Structure of Bacillus subtilis nitric oxide synthase in complex with 6-(4-(((3-Fluorophenethyl)amino)methyl)phenyl)-4-methylpyridin-2- amine | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, 6-[4-({[2-(3-fluorophenyl)ethyl]amino}methyl)phenyl]-4-methylpyridin-2-amine, CHLORIDE ION, ... | | Authors: | Holden, J.K, Poulos, T.L. | | Deposit date: | 2014-11-25 | | Release date: | 2015-07-01 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Nitric Oxide Synthase as a Target for Methicillin-Resistant Staphylococcus Aureus.
Chem.Biol., 22, 2015
|
|
5JWC
 
 | | Structure of NDH2 from plasmodium falciparum in complex with RYL-552 | | Descriptor: | 5-fluoro-3-methyl-2-{4-[4-(trifluoromethoxy)benzyl]phenyl}quinolin-4(1H)-one, FLAVIN-ADENINE DINUCLEOTIDE, FRAGMENT OF TRITON X-100, ... | | Authors: | Yu, Y. | | Deposit date: | 2016-05-12 | | Release date: | 2017-03-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.051 Å) | | Cite: | Target Elucidation by Cocrystal Structures of NADH-Ubiquinone Oxidoreductase of Plasmodium falciparum (PfNDH2) with Small Molecule To Eliminate Drug-Resistant Malaria
J. Med. Chem., 60, 2017
|
|
8XIA
 
 | |
3UGC
 
 | | Structural basis of Jak2 inhibition by the type II inhibtor NVP-BBT594 | | Descriptor: | 5-{[6-(acetylamino)pyrimidin-4-yl]oxy}-N-{4-[(4-methylpiperazin-1-yl)methyl]-3-(trifluoromethyl)phenyl}-2,3-dihydro-1H-indole-1-carboxamide, MALONATE ION, Tyrosine-protein kinase JAK2 | | Authors: | Scheufler, C, Tavares, G.A, Manley, P.W, Pissot-Soldermann, C, Kroemer, M. | | Deposit date: | 2011-11-02 | | Release date: | 2012-05-16 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | Modulation of activation-loop phosphorylation by JAK inhibitors is binding mode dependent.
Cancer Discov, 2, 2012
|
|
