5A5Y
 
 | | Crystal structure of AtTTM3 in complex with tripolyphosphate and magnesium ion (form A) | | Descriptor: | 1,2-ETHANEDIOL, MAGNESIUM ION, TRIPHOSPHATE, ... | | Authors: | Martinez, J, Truffault, V, Hothorn, M. | | Deposit date: | 2015-06-23 | | Release date: | 2015-08-05 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Structural Determinants for Substrate Binding and Catalysis in Triphosphate Tunnel Metalloenzymes.
J.Biol.Chem., 290, 2015
|
|
8JI8
 
 | | Crystal Structure of Prophenoloxidase PPO6 chimeric mutant (F215EASNRAIVD224 to G215DGPDSVVR223) from Aedes aegypti | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, COPPER (II) ION, ... | | Authors: | Zhu, X, Zhang, L, Yang, X, Bao, P, Ren, D, Han, Q. | | Deposit date: | 2023-05-26 | | Release date: | 2023-11-01 | | Last modified: | 2025-04-02 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | The Aedes aegypti mosquito evolves two types of prophenoloxidases with diversified functions.
Proc.Natl.Acad.Sci.USA, 122, 2025
|
|
7YX8
 
 | | Crystal structure of the AM0627 (E326A) inactive mutant in complex with PSGL-1-like bis-T glycopeptide and Zn2+ | | Descriptor: | GLYCEROL, PSGL-1-like bis-T glycopeptide, Peptidase M60 domain-containing protein, ... | | Authors: | Taleb, V, Liao, Q, Narimatsu, Y, Garcia-Garcia, A, Companon, I, Borges, R.J, Gonzalez-Ramirez, A.M, Corzana, F, Clausen, H, Rovira, C, Hurtado-Guerrero, R. | | Deposit date: | 2022-02-15 | | Release date: | 2022-07-20 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural and mechanistic insights into the cleavage of clustered O-glycan patches-containing glycoproteins by mucinases of the human gut.
Nat Commun, 13, 2022
|
|
8Q77
 
 | | STRUCTURE OF PROTEIN KINASE CK2 CATALYTIC SUBUNIT (ISOFORM CK2ALPHA'; CSNK2A2 GENE PRODUCT) IN COMPLEX WITH THE BISUBSTRATE INHIBITOR ARC-780 | | Descriptor: | (2~{S})-2-[[(2~{S})-2-[[(2~{S})-2-[12-[[4-[[5-(4-carboxyphenyl)-1,3-thiazol-2-yl]amino]-4-oxidanylidene-butanoyl]-(2-hydroxy-2-oxoethyl)amino]dodecanoylamino]-4-oxidanyl-4-oxidanylidene-butanoyl]amino]-4-oxidanyl-4-oxidanylidene-butanoyl]amino]butanedioic acid, 1,2-ETHANEDIOL, CHLORIDE ION, ... | | Authors: | Werner, C, Lindenblatt, D, Niefind, K. | | Deposit date: | 2023-08-15 | | Release date: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.255 Å) | | Cite: | Discovery and Exploration of Protein Kinase CK2 Binding Sites Using CK2alpha Cys336Ser as an Exquisite Crystallographic Tool
Kinases Phosphatases, 2023
|
|
5MMR
 
 | | Crystal Structure of CK2alpha with N-((2-chloro-[1,1'-biphenyl]-4-yl)methyl)butane-1,4-diamine bound | | Descriptor: | ACETATE ION, Casein kinase II subunit alpha, PHOSPHATE ION, ... | | Authors: | Brear, P, De Fusco, C, Georgiou, K, Iegre, J, Sore, H, Hyvonen, M, Spring, D. | | Deposit date: | 2016-12-12 | | Release date: | 2017-05-24 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A fragment-based approach leading to the discovery of a novel binding site and the selective CK2 inhibitor CAM4066.
Bioorg. Med. Chem., 25, 2017
|
|
6EKD
 
 | | Crystal structure of JNK3 in complex with a pyridinylimidazole inhibitor | | Descriptor: | 1,2-ETHANEDIOL, 4-(4-methyl-2-methylsulfanyl-1~{H}-imidazol-5-yl)-~{N}-(4-morpholin-4-ylphenyl)pyridin-2-amine, BETA-MERCAPTOETHANOL, ... | | Authors: | Macedo, J.T, Stehle, T, Blaum, B.S. | | Deposit date: | 2017-09-26 | | Release date: | 2018-08-08 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Optimization of a Pyridinylimidazole Scaffold: Shifting the Selectivity from p38 alpha Mitogen-Activated Protein Kinase to c-Jun N-Terminal Kinase 3.
ACS Omega, 3, 2018
|
|
5A60
 
 | | Crystal structure of full-length E. coli ygiF in complex with tripolyphosphate and two magnesium ions | | Descriptor: | 1,2-ETHANEDIOL, INORGANIC TRIPHOSPHATASE, MAGNESIUM ION, ... | | Authors: | Martinez, J, Truffault, V, Hothorn, M. | | Deposit date: | 2015-06-23 | | Release date: | 2015-08-05 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Structural Determinants for Substrate Binding and Catalysis in Triphosphate Tunnel Metalloenzymes.
J.Biol.Chem., 290, 2015
|
|
6G0I
 
 | | Active Fe-PP1 | | Descriptor: | FE (III) ION, MANGANESE (II) ION, PHOSPHATE ION, ... | | Authors: | Salvi, F, Barabas, O, Koehn, M. | | Deposit date: | 2018-03-18 | | Release date: | 2018-11-21 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Effects of stably incorporated iron on protein phosphatase-1 structure and activity.
FEBS Lett., 592, 2018
|
|
8IBQ
 
 | | Bromodomain and Extra-terminal Domain (BET) BRD4 | | Descriptor: | 7-[2-fluoranyl-3-(1,3,5-trimethylpyrazol-4-yl)phenyl]-1~{H}-imidazo[4,5-b]pyridine, Bromodomain-containing protein 4 | | Authors: | Cao, D, Zhiyan, D, Xiong, B. | | Deposit date: | 2023-02-10 | | Release date: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Discovery of 1 H -Imidazo[4,5- b ]pyridine Derivatives as Potent and Selective BET Inhibitors for the Management of Neuropathic Pain.
J.Med.Chem., 66, 2023
|
|
3JST
 
 | |
6GAX
 
 | |
9DHB
 
 | | STRUCTURE OF SERINE HYDROXYMETHYLTRANSFERASE 5 FROM GLYCINE MAX CULTIVAR ESSEX COMPLEXED WITH PLP-GLYCINE AND 5-FORMYLTETRAHYDROFOLATE | | Descriptor: | 1,2-ETHANEDIOL, N-GLYCINE-[3-HYDROXY-2-METHYL-5-PHOSPHONOOXYMETHYL-PYRIDIN-4-YL-METHANE], N-[4-({[(6S)-2-amino-5-formyl-4-oxo-3,4,5,6,7,8-hexahydropteridin-6-yl]methyl}amino)benzoyl]-L-glutamic acid, ... | | Authors: | Beamer, L.J, Owuocha, L.F. | | Deposit date: | 2024-09-03 | | Release date: | 2025-09-17 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | STRUCTURE OF SERINE HYDROXYMETHYLTRANSFERASE 5 FROM GLYCINE MAX CULTIVAR ESSEX COMPLEXED WITH PLP-GLYCINE AND 5-FORMYLTETRAHYDROFOLATE
To Be Published
|
|
8S76
 
 | | Soluble epoxide hydrolase in complex with PROTAC JSF67 | | Descriptor: | 1,2-ETHANEDIOL, Bifunctional epoxide hydrolase 2, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Kumar, A, Schoenfeld, J, Hiesinger, K, Lillich, F, Proschak, E, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2024-02-29 | | Release date: | 2025-03-12 | | Last modified: | 2025-09-24 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Structure-Based Design of PROTACS for the Degradation of Soluble Epoxide Hydrolase.
J.Med.Chem., 68, 2025
|
|
9QFU
 
 | | Human Tryptase beta-2 (hTPSB2) complexed with covalent inhibitor Compound #1 | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Porta, A, Manelfi, C, Talarico, C, Beccari, A.R, Brindisi, M, Summa, V, Iaconis, D, Gobbi, M, Beeg, M. | | Deposit date: | 2025-03-12 | | Release date: | 2025-03-26 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (1.983 Å) | | Cite: | Integrating Surface Plasmon Resonance and Docking Analysis for Mechanistic Insights of Tryptase Inhibitors.
Molecules, 30, 2025
|
|
3K85
 
 | |
8Q7S
 
 | | Crystal structure of the SARS-CoV-2 RBD (Wuhan) with neutralizing VHHs Ma6F06 and Re21H01 | | Descriptor: | 1,2-ETHANEDIOL, 1-ETHOXY-2-(2-ETHOXYETHOXY)ETHANE, GLYCEROL, ... | | Authors: | Guttler, T, Aksu, M, Gorlich, D. | | Deposit date: | 2023-08-16 | | Release date: | 2023-12-20 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Nanobodies to multiple spike variants and inhalation of nanobody-containing aerosols neutralize SARS-CoV-2 in cell culture and hamsters.
Antiviral Res., 221, 2023
|
|
6G3O
 
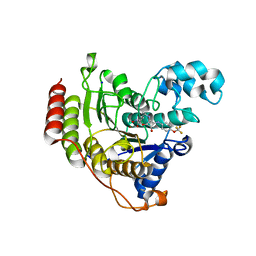 | | Crystal structure of human HDAC2 in complex with (R)-6-[3,4-Dioxo-2-(4-trifluoromethoxy-phenylamino)-cyclobut-1-enylamino]-heptanoic acid hydroxyamide | | Descriptor: | (6~{R})-6-[[3,4-bis(oxidanylidene)-2-[[4-(trifluoromethyloxy)phenyl]amino]cyclobuten-1-yl]amino]-~{N}-oxidanyl-heptanamide, CALCIUM ION, Histone deacetylase 2, ... | | Authors: | Isabet, T, Aurelly, M, Chantalat, L, Thoreau, E. | | Deposit date: | 2018-03-26 | | Release date: | 2018-06-27 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Squaramides as novel class I and IIB histone deacetylase inhibitors for topical treatment of cutaneous t-cell lymphoma.
Bioorg. Med. Chem. Lett., 28, 2018
|
|
6GLG
 
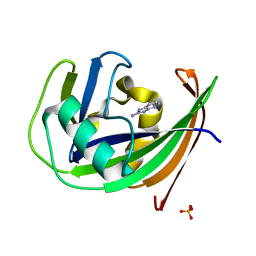 | | Crystal structure of hMTH1 F27A in complex with LW14 in the presence of acetate | | Descriptor: | 1~{H}-imidazo[4,5-b]pyridin-2-amine, 7,8-dihydro-8-oxoguanine triphosphatase, ACETATE ION, ... | | Authors: | Eberle, S.A, Wiedmer, L, Sledz, P, Caflisch, A. | | Deposit date: | 2018-05-23 | | Release date: | 2019-02-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.313 Å) | | Cite: | hMTH1 F27A in complex with LW14
To Be Published
|
|
6RJF
 
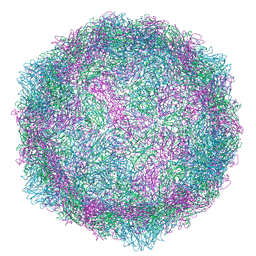 | | Echovirus 1 intact particle | | Descriptor: | PALMITIC ACID, VP1, VP2, ... | | Authors: | Domanska, A, Ruokolainen, V.P, Pelliccia, M, Laajala, M.A, Marjomaki, V.S, Butcher, S.J. | | Deposit date: | 2019-04-26 | | Release date: | 2019-06-12 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Extracellular Albumin and Endosomal Ions Prime Enterovirus Particles for Uncoating That Can Be Prevented by Fatty Acid Saturation.
J.Virol., 93, 2019
|
|
9F0G
 
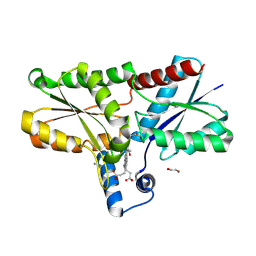 | | LmCpfC H182A variant in complex with iron coproporhyrin III | | Descriptor: | 1,2-ETHANEDIOL, 1,3,5,8-TETRAMETHYL-PORPHINE-2,4,6,7-TETRAPROPIONIC ACID FERROUS COMPLEX, CALCIUM ION, ... | | Authors: | Gabler, T, Hofbauer, S. | | Deposit date: | 2024-04-16 | | Release date: | 2024-04-24 | | Last modified: | 2025-01-15 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Entrance channels to coproheme in coproporphyrin ferrochelatase probed by exogenous imidazole binding.
J.Inorg.Biochem., 260, 2024
|
|
6GLS
 
 | | Crystal structure of hMTH1 N33G in complex with TH scaffold 1 in the absence of acetate | | Descriptor: | 4-phenylpyrimidin-2-amine, 7,8-dihydro-8-oxoguanine triphosphatase, SULFATE ION | | Authors: | Eberle, S.A, Wiedmer, L, Sledz, P, Caflisch, A. | | Deposit date: | 2018-05-23 | | Release date: | 2019-02-20 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.501 Å) | | Cite: | hMTH1 N33G in complex with TH scaffold 1.
To Be Published
|
|
9F0F
 
 | |
6RKF
 
 | | Structure of human DASPO | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, D-aspartate oxidase, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Chaves-Sanjuan, A, Nardini, M. | | Deposit date: | 2019-04-30 | | Release date: | 2020-03-11 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.219 Å) | | Cite: | Structure and kinetic properties of human d-aspartate oxidase, the enzyme-controlling d-aspartate levels in brain.
Faseb J., 34, 2020
|
|
9EUN
 
 | | SARS-CoV-2 nsp10-16 methyltransferase in complex with SAM and m7GTP | | Descriptor: | 1,2-ETHANEDIOL, 2'-O-methyltransferase nsp16, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ... | | Authors: | Kremling, V, Sprenger, J, Oberthuer, D, Scheer, T.E.S. | | Deposit date: | 2024-03-27 | | Release date: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | SARS-CoV-2 methyltransferase nsp10-16 in complex with natural and drug-like purine analogs for guiding structure-based drug development
To Be Published
|
|
9EMJ
 
 | | SARS-CoV-2 methyltransferase nsp10-16 in complex with Toyocamycin and m7GpppA (Cap0-analog) | | Descriptor: | 1,2-ETHANEDIOL, 2'-O-methyltransferase nsp16, 4-amino-7-(beta-D-ribofuranosyl)-7H-pyrrolo[2,3-d]pyrimidine-5-carbonitrile, ... | | Authors: | Kremling, V, Sprenger, J, Oberthuer, D, Kiene, A. | | Deposit date: | 2024-03-08 | | Release date: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | SARS-CoV-2 methyltransferase nsp10-16 in complex with natural and drug-like purine analogs for guiding structure-based drug development
To Be Published
|
|
