5A5Y
 
 | | Crystal structure of AtTTM3 in complex with tripolyphosphate and magnesium ion (form A) | | Descriptor: | 1,2-ETHANEDIOL, MAGNESIUM ION, TRIPHOSPHATE, ... | | Authors: | Martinez, J, Truffault, V, Hothorn, M. | | Deposit date: | 2015-06-23 | | Release date: | 2015-08-05 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Structural Determinants for Substrate Binding and Catalysis in Triphosphate Tunnel Metalloenzymes.
J.Biol.Chem., 290, 2015
|
|
5D09
 
 | |
1E66
 
 | | STRUCTURE OF ACETYLCHOLINESTERASE COMPLEXED WITH (-)-HUPRINE X AT 2.1A RESOLUTION | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 3-CHLORO-9-ETHYL-6,7,8,9,10,11-HEXAHYDRO-7,11-METHANOCYCLOOCTA[B]QUINOLIN-12-AMINE, ACETYLCHOLINESTERASE | | Authors: | Dvir, H, Harel, M, Silman, I, Sussman, J.L. | | Deposit date: | 2000-08-08 | | Release date: | 2001-08-02 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | 3D Structure of Torpedo Californica Acetylcholinesterase Complexed with Huprine X at 2. 1 A Resolution: Kinetic and Molecular Dynamic Correlates.
Biochemistry, 41, 2002
|
|
307D
 
 | | Structure of a DNA analog of the primer for HIV-1 RT second strand synthesis | | Descriptor: | DNA (5'-D(*CP*AP*AP*AP*GP*AP*AP*AP*AP*G)-3'), DNA (5'-D(*CP*TP*TP*TP*TP*CP*TP*TP*TP*G)-3') | | Authors: | Han, G.W, Kopka, M.L, Cascio, D, Grzeskowiak, K, Dickerson, R.E. | | Deposit date: | 1997-01-07 | | Release date: | 1997-01-27 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure of a DNA analog of the primer for HIV-1 RT second strand synthesis.
J.Mol.Biol., 269, 1997
|
|
8XVJ
 
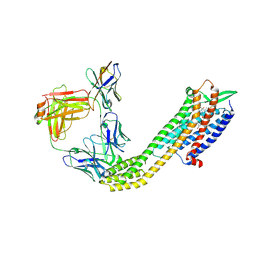 | | Cryo-EM structure of ETAR bound with Macitentan | | Descriptor: | Endoglucanase H,Endothelin-1 receptor,Soluble cytochrome b562, Macitentan, anti-BRIL Fab Heavy chain, ... | | Authors: | Hou, J.Y, Liu, S.H, Wu, L.J, Liu, Z.J, Hua, T. | | Deposit date: | 2024-01-15 | | Release date: | 2024-08-28 | | Last modified: | 2025-07-23 | | Method: | ELECTRON MICROSCOPY (3.26 Å) | | Cite: | Structural basis of antagonist selectivity in endothelin receptors.
Cell Discov, 10, 2024
|
|
8XVK
 
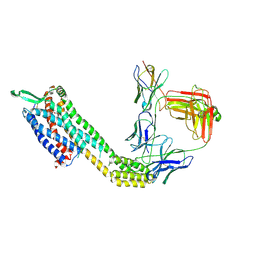 | | Cryo-EM structure of ETAR bound with Ambrisentan | | Descriptor: | Ambrisentan, Endoglucanase H,Endothelin-1 receptor,Soluble cytochrome b562, anti-BRIL Fab Heavy chain, ... | | Authors: | Hou, J.Y, Liu, S.H, Wu, L.J, Liu, Z.J, Hua, T. | | Deposit date: | 2024-01-15 | | Release date: | 2024-08-28 | | Last modified: | 2025-07-23 | | Method: | ELECTRON MICROSCOPY (3.21 Å) | | Cite: | Structural basis of antagonist selectivity in endothelin receptors.
Cell Discov, 10, 2024
|
|
5NL5
 
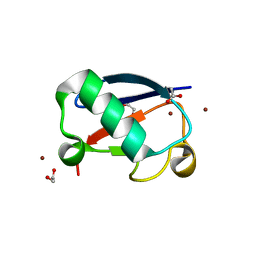 | | Crystal structure of Zn1.7-E16V human ubiquitin (hUb) mutant adduct, from a solution 70 mM zinc acetate/1.3 mM E16V hUb | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Polyubiquitin-B, ... | | Authors: | Fermani, S, Falini, G. | | Deposit date: | 2017-04-04 | | Release date: | 2017-04-26 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Aggregation Pathways of Native-Like Ubiquitin Promoted by Single-Point Mutation, Metal Ion Concentration, and Dielectric Constant of the Medium.
Chemistry, 24, 2018
|
|
8XVL
 
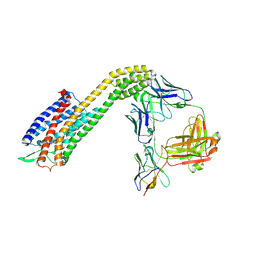 | | Cryo-EM structure of ETAR bound with Zibotentan | | Descriptor: | Endoglucanase H,Endothelin-1 receptor,Soluble cytochrome b562, Zibotentan, anti-BRIL Fab Heavy chain, ... | | Authors: | Hou, J.Y, Liu, S.H, Wu, L.J, Liu, Z.J, Hua, T. | | Deposit date: | 2024-01-15 | | Release date: | 2024-08-28 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3.22 Å) | | Cite: | Structural basis of antagonist selectivity in endothelin receptors.
Cell Discov, 10, 2024
|
|
7TI0
 
 | | Structure of CTX-M-15 bound to RPX-7063 at 1.5A | | Descriptor: | 1,2-ETHANEDIOL, Beta-lactamase, CHLORIDE ION, ... | | Authors: | Clifton, M.C, Abendroth, J, Hecker, S.J, Edwards, T.E. | | Deposit date: | 2022-01-12 | | Release date: | 2022-05-18 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Broad-spectrum cyclic boronate beta-lactamase inhibitors featuring an intramolecular prodrug for oral bioavailability.
Bioorg.Med.Chem., 62, 2022
|
|
5NLJ
 
 | | Crystal structure of Zn3-E16V human ubiquitin (hUb) mutant adduct, from a solution 70 mM zinc acetate/20% v/v TFE/1.3 mM E16V hUb | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Fermani, S, Falini, G. | | Deposit date: | 2017-04-04 | | Release date: | 2017-04-26 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Aggregation Pathways of Native-Like Ubiquitin Promoted by Single-Point Mutation, Metal Ion Concentration, and Dielectric Constant of the Medium.
Chemistry, 24, 2018
|
|
6WDP
 
 | | Interleukin 12 receptor subunit beta-1 | | Descriptor: | GLYCEROL, Interleukin-12 receptor subunit beta-1, SULFATE ION | | Authors: | Spangler, J.B, Thomas, C, Jude, K.M, Garcia, K.C. | | Deposit date: | 2020-04-01 | | Release date: | 2021-02-24 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Structural basis for IL-12 and IL-23 receptor sharing reveals a gateway for shaping actions on T versus NK cells.
Cell, 184, 2021
|
|
7S49
 
 | | Crystal Structure of Inhibitor-bound Galactokinase | | Descriptor: | (4R)-2-[(1,3-benzoxazol-2-yl)amino]-4-(4-chloro-1H-pyrazol-5-yl)-4,6,7,8-tetrahydroquinazolin-5(1H)-one, Galactokinase, PHOSPHATE ION, ... | | Authors: | Whitby, F.G. | | Deposit date: | 2021-09-08 | | Release date: | 2021-09-29 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure-Based Optimization of Small Molecule Human Galactokinase Inhibitors.
J.Med.Chem., 64, 2021
|
|
8PK0
 
 | | human mitoribosomal large subunit assembly intermediate 1 with GTPBP10-GTPBP7 | | Descriptor: | 16S rRNA + pre-H68-71 segment, 39S ribosomal protein L10, mitochondrial, ... | | Authors: | Kummer, E, Nguyen, T.G, Ritter, C. | | Deposit date: | 2023-06-23 | | Release date: | 2023-12-13 | | Last modified: | 2025-10-01 | | Method: | ELECTRON MICROSCOPY (3.03 Å) | | Cite: | Structural insights into the role of GTPBP10 in the RNA maturation of the mitoribosome.
Nat Commun, 14, 2023
|
|
7YD3
 
 | | Single-chain variable fragment of app 3D1 antibody | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Yan, L, Yang, G. | | Deposit date: | 2022-07-03 | | Release date: | 2023-07-05 | | Last modified: | 2025-08-27 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | A broadly neutralizing antibody recognizes a unique epitope with a signature motif common across coronaviruses
Nat Commun, 16, 2025
|
|
8PQI
 
 | | PDGFRA T674I mutant kinase domain in complex with avapritinib derivative 9 | | Descriptor: | (1~{S})-~{N}-ethyl-1-(4-fluorophenyl)-1-[2-[4-[6-(1-methylpyrazol-4-yl)pyrrolo[2,1-f][1,2,4]triazin-4-yl]piperazin-1-yl]pyrimidin-5-yl]ethanamine, DI(HYDROXYETHYL)ETHER, Platelet-derived growth factor receptor alpha | | Authors: | Teuber, A, Kleinboelting, S, Mueller, M.P, Rauh, D. | | Deposit date: | 2023-07-11 | | Release date: | 2023-12-27 | | Last modified: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Avapritinib-based SAR studies unveil a binding pocket in KIT and PDGFRA.
Nat Commun, 15, 2024
|
|
8XET
 
 | | High-resolution structure of the siderophore periplasmic binding protein FtsB from Streptococcus pyogenes | | Descriptor: | 1,2-ETHANEDIOL, 3,6,9,12,15,18-HEXAOXAICOSANE-1,20-DIOL, CHLORIDE ION, ... | | Authors: | Caaveiro, J.M.M, Fernandez-Perez, J, Tsumoto, K. | | Deposit date: | 2023-12-13 | | Release date: | 2024-10-09 | | Last modified: | 2024-12-18 | | Method: | X-RAY DIFFRACTION (1.11 Å) | | Cite: | Structural basis for the ligand promiscuity of the hydroxamate siderophore binding protein FtsB from Streptococcus pyogenes.
Structure, 32, 2024
|
|
6ULJ
 
 | | Crystal structure of human GAC in complex with inhibitor UPGL00012 | | Descriptor: | 2-phenyl-N-{6-[4-({6-[(phenylacetyl)amino]pyridazin-3-yl}oxy)piperidin-1-yl]pyridazin-3-yl}acetamide, Glutaminase kidney isoform, mitochondrial | | Authors: | Huang, Q, Cerione, R.A. | | Deposit date: | 2019-10-08 | | Release date: | 2020-10-14 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Crystal structure of human GAC in complex with inhibitor UPGL00045
To Be Published
|
|
5O12
 
 | |
3IQD
 
 | | Structure of Octopine-dehydrogenase in complex with NADH and Agmatine | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, AGMATINE, Octopine dehydrogenase | | Authors: | Smits, S.H.J, Meyer, T, Mueller, A, Willbold, D, Grieshaber, M.K, Schmitt, L. | | Deposit date: | 2009-08-20 | | Release date: | 2010-08-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Insights into the mechanism of ligand binding to octopine dehydrogenase from Pecten maximus by NMR and crystallography
Plos One, 5, 2010
|
|
5KMJ
 
 | | TrkA JM-kinase with {N}-(2-pyridylmethyl)-2-[2-(2-thienyl)indol-1-yl]acetamide | | Descriptor: | High affinity nerve growth factor receptor, ~{N}-(pyridin-2-ylmethyl)-2-(2-thiophen-2-ylindol-1-yl)ethanamide | | Authors: | Su, H.P. | | Deposit date: | 2016-06-27 | | Release date: | 2016-12-28 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Structural characterization of nonactive site, TrkA-selective kinase inhibitors.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
7N7R
 
 | | Crystal Structure of SARS-CoV-2 NendoU in complex with Z2472938267 | | Descriptor: | 1-[2-(2-oxidanylidenepyrrolidin-1-yl)ethyl]-3-phenyl-urea, Uridylate-specific endoribonuclease | | Authors: | Godoy, A.S, Douangamath, A, Nakamura, A.M, Dias, A, Krojer, T, Noske, G.D, Gawiljuk, V.O, Fernandes, R.S, Fairhead, M, Powell, A, Dunnet, L, Aimon, A, Fearon, D, Brandao-Neto, J, Skyner, R, von Delft, F, Oliva, G. | | Deposit date: | 2021-06-11 | | Release date: | 2021-09-08 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Allosteric regulation and crystallographic fragment screening of SARS-CoV-2 NSP15 endoribonuclease.
Nucleic Acids Res., 51, 2023
|
|
6ISE
 
 | | Crystal structure of AMPPNP bound CK2 alpha from C. neoformans | | Descriptor: | 1,2-ETHANEDIOL, Casein kinase II subunit alpha, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Cho, H.S. | | Deposit date: | 2018-11-16 | | Release date: | 2019-11-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of AMPPNP bound CK2 alpha from C. neoformans
To be published
|
|
8R4V
 
 | | Structure of Salt-inducible kinase 3 in complex with inhibitor | | Descriptor: | 1-(2,4-dimethoxyphenyl)-3-(2,6-dimethylphenyl)-1-[6-[[4-(4-methylpiperazin-1-yl)phenyl]amino]pyrimidin-4-yl]urea, Serine/threonine-protein kinase SIK3 | | Authors: | Kack, H, Oster, L. | | Deposit date: | 2023-11-14 | | Release date: | 2024-03-27 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The structures of salt-inducible kinase 3 in complex with inhibitors reveal determinants for binding and selectivity.
J.Biol.Chem., 300, 2024
|
|
6BTY
 
 | | Crystal structure of the PI3KC2alpha C2 domain in space group P41212 | | Descriptor: | 1,4,7,10,13,16-HEXAOXACYCLOOCTADECANE, Phosphatidylinositol 4-phosphate 3-kinase C2 domain-containing subunit alpha | | Authors: | Chen, K.-E, Collins, B.M. | | Deposit date: | 2017-12-08 | | Release date: | 2018-10-17 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.678 Å) | | Cite: | Molecular Basis for Membrane Recruitment by the PX and C2 Domains of Class II Phosphoinositide 3-Kinase-C2 alpha.
Structure, 26, 2018
|
|
9BR7
 
 | | Crystal structure of human succinyl-CoA:glutarate-CoA transferase (SUGCT) in complex with Losartan carboxylic acid | | Descriptor: | AMMONIUM ION, SULFATE ION, Succinate--hydroxymethylglutarate CoA-transferase, ... | | Authors: | Wu, R, Lazarus, M.B. | | Deposit date: | 2024-05-10 | | Release date: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Characterization, Structure, and Inhibition of the Human Succinyl-CoA:glutarate-CoA Transferase, a Putative Genetic Modifier of Glutaric Aciduria Type 1.
Acs Chem.Biol., 19, 2024
|
|
